-
如何利用DGL官方库中的rgcn链接预测代码跑自己的数据集(如何在DGL库的链接预测数据集模块定义自己的数据集类)
最近在忙我的省创,是有关于知识图谱的,其中有一个内容是使用rgcn的链接预测方法跑自己的数据集,我是用的dgl库中给出的在pytorch环境下实现rgcn的链接预测的代码,相关链接贴在这里:
dgl库中在pytorch环境下实现rgcn的链接预测的代码
这个代码给的示例就是使用FB15k237数据集,调用方法是这样的:
from dgl.data.knowledge_graph import FB15k237Dataset data = FB15k237Dataset(reverse=False) graph = data[0] print("graph",graph)- 1
- 2
- 3
- 4
这里就调用了FB15k237数据集,返回的的
data[0]就是使用dgl库使用该数据集构建的图g。我一开始想用自己的数据构图,然后使用rgcn的代码跑我自己的数据集,但是我不知道它的构图是如何实现的,于是我修改了rgcn的代码,实现了自己的构图方式如下,就是使用入结点出节点和边的编号列表构图:
g = dgl.graph((src, dst), num_nodes=num_nodes) g.edata[dgl.ETYPE] = rel- 1
- 2
鉴于rgcn示例里使用的FB15k237数据集的图的属性有
'train_mask'和'test_mask'等属性,我就把rgcn代码里有关构图的部分全改成我自己的了,修改过后的完整可运行rgcn代码如下。这个代码需要自己提供
entity.txt,relation.txt,train.txt,valid.txt,test.txt五个文件,entity.txt和relation.txt分别代表实体编号到实体描述的映射,关系编号到关系描述的映射,类似这样:
train.txt,valid.txt,test.txt这三个文件就代表训练集,验证集和测试集的已经被映射为编号的(h,r,t)格式的三元组,类似这样:
在代码中写入对应的自己的数据集已经处理好的这五个文件的地址,运行下面的文件就可以运行完整的rgcn代码了:import numpy as np import torch import torch.nn as nn import scipy as sp import torch.nn.functional as F import dgl from dgl.data.knowledge_graph import FB15k237Dataset from dgl.data.knowledge_graph import FB15kDataset from dgl.dataloading import GraphDataLoader from dgl.nn.pytorch import RelGraphConv import tqdm # for building training/testing graphs def get_subset_g(g, mask, num_rels, bidirected=False): src, dst = g.edges() sub_src = src[mask] sub_dst = dst[mask] sub_rel = g.edata['etype'][mask] if bidirected: sub_src, sub_dst = torch.cat([sub_src, sub_dst]), torch.cat([sub_dst, sub_src]) sub_rel = torch.cat([sub_rel, sub_rel + num_rels]) sub_g = dgl.graph((sub_src, sub_dst), num_nodes=g.num_nodes()) sub_g.edata[dgl.ETYPE] = sub_rel return sub_g class GlobalUniform: def __init__(self, g, sample_size): self.sample_size = sample_size self.eids = np.arange(g.num_edges(),dtype='int64') def sample(self): return torch.from_numpy(np.random.choice(self.eids, self.sample_size)) class NegativeSampler: def __init__(self, k=10): # negative sampling rate = 10 self.k = k def sample(self, pos_samples, num_nodes): batch_size = len(pos_samples) neg_batch_size = batch_size * self.k neg_samples = np.tile(pos_samples, (self.k, 1)) values = np.random.randint(num_nodes, size=neg_batch_size) choices = np.random.uniform(size=neg_batch_size) subj = choices > 0.5 obj = choices <= 0.5 neg_samples[subj, 0] = values[subj] neg_samples[obj, 2] = values[obj] samples = np.concatenate((pos_samples, neg_samples)) # binary labels indicating positive and negative samples labels = np.zeros(batch_size * (self.k + 1), dtype=np.float32) labels[:batch_size] = 1 return torch.from_numpy(samples), torch.from_numpy(labels) class SubgraphIterator: def __init__(self, g, num_rels, sample_size=30000, num_epochs=6000): self.g = g self.num_rels = num_rels self.sample_size = sample_size self.num_epochs = num_epochs self.pos_sampler = GlobalUniform(g, sample_size) self.neg_sampler = NegativeSampler() def __len__(self): return self.num_epochs def __getitem__(self, i): eids = self.pos_sampler.sample() src, dst = self.g.find_edges(eids) src, dst = src.numpy(), dst.numpy() rel = self.g.edata[dgl.ETYPE][eids].numpy() # relabel nodes to have consecutive node IDs uniq_v, edges = np.unique((src, dst), return_inverse=True) num_nodes = len(uniq_v) # edges is the concatenation of src, dst with relabeled ID src, dst = np.reshape(edges, (2, -1)) relabeled_data = np.stack((src, rel, dst)).transpose() samples, labels = self.neg_sampler.sample(relabeled_data, num_nodes) # use only half of the positive edges chosen_ids = np.random.choice(np.arange(self.sample_size), size=int(self.sample_size / 2), replace=False) src = src[chosen_ids] dst = dst[chosen_ids] rel = rel[chosen_ids] src, dst = np.concatenate((src, dst)), np.concatenate((dst, src)) rel = np.concatenate((rel, rel + self.num_rels)) sub_g = dgl.graph((src, dst), num_nodes=num_nodes) sub_g.edata[dgl.ETYPE] = torch.from_numpy(rel) sub_g.edata['norm'] = dgl.norm_by_dst(sub_g).unsqueeze(-1) uniq_v = torch.from_numpy(uniq_v).view(-1).long() return sub_g, uniq_v, samples, labels class RGCN(nn.Module): def __init__(self, num_nodes, h_dim, num_rels): super().__init__() # two-layer RGCN self.emb = nn.Embedding(num_nodes, h_dim) self.conv1 = RelGraphConv(h_dim, h_dim, num_rels, regularizer='bdd', num_bases=100, self_loop=True) self.conv2 = RelGraphConv(h_dim, h_dim, num_rels, regularizer='bdd', num_bases=100, self_loop=True) self.dropout = nn.Dropout(0.2) def forward(self, g, nids): x = self.emb(nids) h = F.relu(self.conv1(g, x, g.edata[dgl.ETYPE], g.edata['norm'])) h = self.dropout(h) h = self.conv2(g, h, g.edata[dgl.ETYPE], g.edata['norm']) return self.dropout(h) class LinkPredict(nn.Module): def __init__(self, num_nodes, num_rels, h_dim = 500, reg_param=0.01): super().__init__() self.rgcn = RGCN(num_nodes, h_dim, num_rels * 2) self.reg_param = reg_param self.w_relation = nn.Parameter(torch.Tensor(num_rels, h_dim)) nn.init.xavier_uniform_(self.w_relation, gain=nn.init.calculate_gain('relu')) def calc_score(self, embedding, triplets): s = embedding[triplets[:,0]] r = self.w_relation[triplets[:,1]] o = embedding[triplets[:,2]] score = torch.sum(s * r * o, dim=1) return score def forward(self, g, nids): return self.rgcn(g, nids) def regularization_loss(self, embedding): return torch.mean(embedding.pow(2)) + torch.mean(self.w_relation.pow(2)) def get_loss(self, embed, triplets, labels): # each row in the triplets is a 3-tuple of (source, relation, destination) score = self.calc_score(embed, triplets) predict_loss = F.binary_cross_entropy_with_logits(score, labels) reg_loss = self.regularization_loss(embed) return predict_loss + self.reg_param * reg_loss def filter(triplets_to_filter, target_s, target_r, target_o, num_nodes, filter_o=True): """Get candidate heads or tails to score""" target_s, target_r, target_o = int(target_s), int(target_r), int(target_o) # Add the ground truth node first if filter_o: candidate_nodes = [target_o] else: candidate_nodes = [target_s] for e in range(num_nodes): triplet = (target_s, target_r, e) if filter_o else (e, target_r, target_o) # Do not consider a node if it leads to a real triplet if triplet not in triplets_to_filter: candidate_nodes.append(e) return torch.LongTensor(candidate_nodes) def perturb_and_get_filtered_rank(emb, w, s, r, o, test_size, triplets_to_filter, filter_o=True): """Perturb subject or object in the triplets""" num_nodes = emb.shape[0] ranks = [] for idx in tqdm.tqdm(range(test_size), desc="Evaluate"): target_s = s[idx] target_r = r[idx] target_o = o[idx] candidate_nodes = filter(triplets_to_filter, target_s, target_r, target_o, num_nodes, filter_o=filter_o) if filter_o: emb_s = emb[target_s] emb_o = emb[candidate_nodes] else: emb_s = emb[candidate_nodes] emb_o = emb[target_o] target_idx = 0 emb_r = w[target_r] emb_triplet = emb_s * emb_r * emb_o scores = torch.sigmoid(torch.sum(emb_triplet, dim=1)) _, indices = torch.sort(scores, descending=True) rank = int((indices == target_idx).nonzero()) ranks.append(rank) return torch.LongTensor(ranks) def calc_mrr(emb, w, triplets_to_filter, batch_size=100, filter=True): with torch.no_grad(): test_triplets = triplets_to_filter s, r, o = test_triplets[:,0], test_triplets[:,1], test_triplets[:,2] test_size = len(s) triplets_to_filter = {tuple(triplet) for triplet in triplets_to_filter.tolist()} ranks_s = perturb_and_get_filtered_rank(emb, w, s, r, o, test_size, triplets_to_filter, filter_o=False) ranks_o = perturb_and_get_filtered_rank(emb, w, s, r, o, test_size, triplets_to_filter) ranks = torch.cat([ranks_s, ranks_o]) ranks += 1 # change to 1-indexed mrr = torch.mean(1.0 / ranks.float()).item() mr = torch.mean(ranks.float()).item() print("MRR (filtered): {:.6f}".format(mrr)) print("MR (filtered): {:.6f}".format(mr)) hits=[1,3,10] for hit in hits: avg_count = torch.mean((ranks <= hit).float()) print("Hits (filtered) @ {}: {:.6f}".format(hit, avg_count.item())) return mrr def train(dataloader, test_g, test_nids, triplets, device, model_state_file, model): optimizer = torch.optim.Adam(model.parameters(), lr=1e-2) best_mrr = 0 for epoch, batch_data in enumerate(dataloader): # single graph batch model.train() g, train_nids, edges, labels = batch_data g = g.to(device) train_nids = train_nids.to(device) edges = edges.to(device) labels = labels.to(device) embed = model(g, train_nids) loss = model.get_loss(embed, edges, labels) optimizer.zero_grad() loss.backward() nn.utils.clip_grad_norm_(model.parameters(), max_norm=1.0) # clip gradients optimizer.step() print("Epoch {:04d} | Loss {:.4f} | Best MRR {:.4f}".format(epoch, loss.item(), best_mrr)) if (epoch + 1) % 500 == 0: # perform validation on CPU because full graph is too large model = model.cpu() model.eval() embed = model(test_g, test_nids) mrr = calc_mrr(embed, model.w_relation, triplets, batch_size=500) # save best model if best_mrr < mrr: best_mrr = mrr torch.save({'state_dict': model.state_dict(), 'epoch': epoch}, model_state_file) model = model.to(device) if __name__ == '__main__': device = torch.device('cuda' if torch.cuda.is_available() else 'cpu') print(f'Training with DGL built-in RGCN module') # load and preprocess dataset # data = FB15k237Dataset(reverse=False) # data = FB15kDataset(reverse=False) entityfile=r'data/entity.txt' relationfile=r'data/relation.txt' f1 = open(entityfile, 'r') f2 = open(relationfile, 'r') entity=[] relation=[] for line in f1: l=line.strip().split("\t") entity.append(int(l[0])) for line in f2: l=line.strip().split("\t") relation.append(int(l[0])) num_nodes=len(entity) num_rels=len(relation) n_entities=num_nodes print("# entities:",num_nodes) print("# relations:",num_rels) trainfile=r'data/train.txt' f3 = open(trainfile, 'r') src_train=[] rel_train=[] dst_train=[] for line in f3: l=line.strip().split("\t") h=int(l[0]) r=int(l[1]) t=int(l[2]) src_train.append(h) rel_train.append(r) dst_train.append(t) print("# training edges: ",len(src_train)) src_train=torch.LongTensor(src_train) rel_train=torch.LongTensor(rel_train) dst_train=torch.LongTensor(dst_train) train_g = dgl.graph((src_train, dst_train), num_nodes=num_nodes) train_g.edata[dgl.ETYPE] = rel_train src_test, dst_test = torch.cat([src_train, dst_train]), torch.cat([dst_train,src_train]) rel_test = torch.cat([rel_train, rel_train + num_rels]) test_g = dgl.graph((src_test, dst_test), num_nodes=num_nodes) test_g.edata[dgl.ETYPE] = rel_test test_g.edata['norm'] = dgl.norm_by_dst(test_g).unsqueeze(-1) test_nids = torch.arange(0, num_nodes) subg_iter = SubgraphIterator(train_g, num_rels) # uniform edge sampling dataloader = GraphDataLoader(subg_iter, batch_size=1, collate_fn=lambda x: x[0]) validfile=r'data/valid.txt' f4 = open(validfile, 'r') num_valid=0 for line in f4: num_valid+=1 print("# validation edges: ",num_valid) # Prepare data for metric computation testfile=r'data/test.txt' f5 = open(testfile, 'r') src=[] rel=[] dst=[] for line in f5: l=line.strip().split("\t") h=int(l[0]) r=int(l[1]) t=int(l[2]) src.append(h) rel.append(r) dst.append(t) print("# testing edges: ",len(src)) src=torch.LongTensor(src) rel=torch.LongTensor(rel) dst=torch.LongTensor(dst) triplets_test = torch.stack([src,rel, dst], dim=1) # create RGCN model model = LinkPredict(num_nodes, num_rels).to(device) # train model_state_file = 'model_state.pth' train(dataloader, test_g, test_nids, triplets_test, device, model_state_file, model) # testing print("Testing...") checkpoint = torch.load(model_state_file) model = model.cpu() # test on CPU model.eval() model.load_state_dict(checkpoint['state_dict']) embed = model(test_g, test_nids) best_mrr = calc_mrr(embed, model.w_relation,triplets_test, batch_size=500) print("Best MRR {:.4f} achieved using the epoch {:04d}".format(best_mrr, checkpoint['epoch']))- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
- 45
- 46
- 47
- 48
- 49
- 50
- 51
- 52
- 53
- 54
- 55
- 56
- 57
- 58
- 59
- 60
- 61
- 62
- 63
- 64
- 65
- 66
- 67
- 68
- 69
- 70
- 71
- 72
- 73
- 74
- 75
- 76
- 77
- 78
- 79
- 80
- 81
- 82
- 83
- 84
- 85
- 86
- 87
- 88
- 89
- 90
- 91
- 92
- 93
- 94
- 95
- 96
- 97
- 98
- 99
- 100
- 101
- 102
- 103
- 104
- 105
- 106
- 107
- 108
- 109
- 110
- 111
- 112
- 113
- 114
- 115
- 116
- 117
- 118
- 119
- 120
- 121
- 122
- 123
- 124
- 125
- 126
- 127
- 128
- 129
- 130
- 131
- 132
- 133
- 134
- 135
- 136
- 137
- 138
- 139
- 140
- 141
- 142
- 143
- 144
- 145
- 146
- 147
- 148
- 149
- 150
- 151
- 152
- 153
- 154
- 155
- 156
- 157
- 158
- 159
- 160
- 161
- 162
- 163
- 164
- 165
- 166
- 167
- 168
- 169
- 170
- 171
- 172
- 173
- 174
- 175
- 176
- 177
- 178
- 179
- 180
- 181
- 182
- 183
- 184
- 185
- 186
- 187
- 188
- 189
- 190
- 191
- 192
- 193
- 194
- 195
- 196
- 197
- 198
- 199
- 200
- 201
- 202
- 203
- 204
- 205
- 206
- 207
- 208
- 209
- 210
- 211
- 212
- 213
- 214
- 215
- 216
- 217
- 218
- 219
- 220
- 221
- 222
- 223
- 224
- 225
- 226
- 227
- 228
- 229
- 230
- 231
- 232
- 233
- 234
- 235
- 236
- 237
- 238
- 239
- 240
- 241
- 242
- 243
- 244
- 245
- 246
- 247
- 248
- 249
- 250
- 251
- 252
- 253
- 254
- 255
- 256
- 257
- 258
- 259
- 260
- 261
- 262
- 263
- 264
- 265
- 266
- 267
- 268
- 269
- 270
- 271
- 272
- 273
- 274
- 275
- 276
- 277
- 278
- 279
- 280
- 281
- 282
- 283
- 284
- 285
- 286
- 287
- 288
- 289
- 290
- 291
- 292
- 293
- 294
- 295
- 296
- 297
- 298
- 299
- 300
- 301
- 302
- 303
- 304
- 305
- 306
- 307
- 308
- 309
- 310
- 311
- 312
- 313
- 314
- 315
- 316
- 317
- 318
- 319
- 320
- 321
- 322
- 323
- 324
- 325
- 326
- 327
- 328
- 329
- 330
- 331
- 332
- 333
- 334
- 335
- 336
- 337
- 338
- 339
但是,这个代码的效果并不太好,贴在这里只是做个过程记录,同样的数据集,为什么这样简单的构图效果就没有dgl库里自己构图的效果好呢?说实话我也不知道(°ー°〃)我也看了dgl库里处理数据然后构图的代码,确实要精细很多,我就认为是预处理数据的方式不一样导致效果的差别吧。因此下面要说的就是如何在如何在DGL库的链接预测数据集模块定义自己的数据集类,将自己的数据集输入,使用dgl库中处理数据的方法处理我们的数据,再像刚刚调用FB15k237数据集那样调用自己的数据集。
- step 1 :
找到你的
dgl.data.knowledge_graph.py文件,(我这里使用的版本是dgl 0.9.0),在这个文件中,定义了FB15k237Dataset,FB15Dataset和WN18Dataset三个常用的知识图谱数据集类,我们添加一个自己的数据集类MyDataset(其实就是copy了一下别的类(°ー°〃))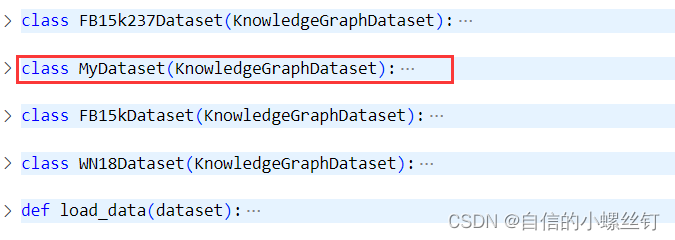
把name改成mydata:class MyDataset(KnowledgeGraphDataset): def __init__(self, reverse=True, raw_dir=None, force_reload=False, verbose=True, transform=None): name = 'mydata' super(MyDataset, self).__init__(name, reverse, raw_dir, force_reload, verbose, transform) def __getitem__(self, idx): r"""Gets the graph object """ return super(MyDataset, self).__getitem__(idx) def __len__(self): r"""The number of graphs in the dataset.""" return super(MyDataset, self).__len__()- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- step 2:
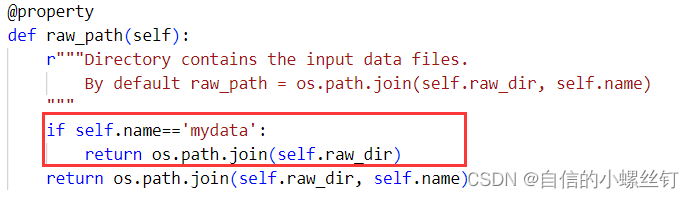
找到你的
dgl.data.dgl_dataset.py文件,找到下图对应的代码位置,加入框框内的代码:
(至于为什么要这样呢,,,,自己看代码吧,虽然我也很想做记录,方便自己下次看懂,但是感觉要讲的话将不太清楚,打半天字解释不如自己看看代码咋写的 ┭┮﹏┭┮)if self.name=='mydata': return os.path.join(self.raw_dir)- 1
- 2
- step 3:
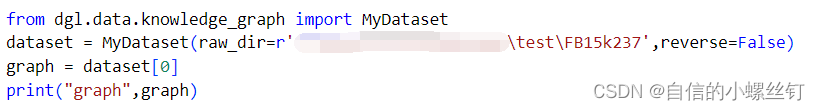
在rgcn的链接预测代码里调用一下自己的数据就好啦,下面是一个简单的
demo,这样就可以调用自己的数据集类了。from dgl.data.knowledge_graph import MyDataset dataset = MyDataset(raw_dir=r'你自己装数据集的文件夹位置',reverse=False)- 1
- 2
- step 4:还有十分重要的一点就是,数据集的格式,我是把自己的数据集都设成了和它调用的FB15k237数据集一样的格式,因为
step 3中要写入的文件夹地址内要包含的文件有5个:entities.dict,relations.dict,train.txt,valid.txt,test.txt。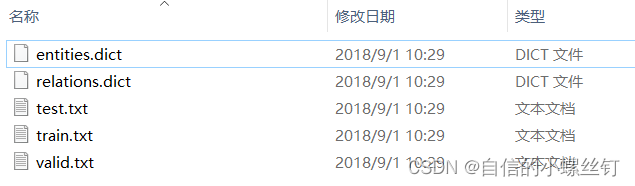
entities.dict和relations.dict分别代表实体编号到实体描述的映射,关系编号到关系描述的映射,类似这样:
train.txt,valid.txt,test.txt这三个文件代表训练集,验证集和测试集的还没有被映射为编号的(h,r,t)格式的三元组,类似这样:(它们中间的间隔均是'\t')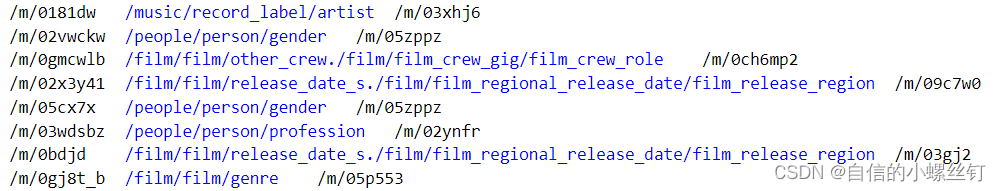
把我改过的最终的rgcn代码贴在下面,做个记录,其中我对
calc_mrr函数做了修改的,它原本的代码里只有mrr一个评估指标,我增加了mr,hist@1,hist@3,hist@10这几个指标,在代码里看吧:import numpy as np import torch import torch.nn as nn import torch.nn.functional as F import dgl from dgl.data.knowledge_graph import FB15k237Dataset from dgl.data.knowledge_graph import FB15kDataset from dgl.data.knowledge_graph import MyDataset from dgl.dataloading import GraphDataLoader from dgl.nn.pytorch import RelGraphConv import tqdm # for building training/testing graphs def get_subset_g(g, mask, num_rels, bidirected=False): src, dst = g.edges() sub_src = src[mask] sub_dst = dst[mask] sub_rel = g.edata['etype'][mask] if bidirected: sub_src, sub_dst = torch.cat([sub_src, sub_dst]), torch.cat([sub_dst, sub_src]) sub_rel = torch.cat([sub_rel, sub_rel + num_rels]) sub_g = dgl.graph((sub_src, sub_dst), num_nodes=g.num_nodes()) sub_g.edata[dgl.ETYPE] = sub_rel return sub_g class GlobalUniform: def __init__(self, g, sample_size): self.sample_size = sample_size self.eids = np.arange(g.num_edges()) def sample(self): return torch.from_numpy(np.random.choice(self.eids, self.sample_size)) class NegativeSampler: def __init__(self, k=10): # negative sampling rate = 10 self.k = k def sample(self, pos_samples, num_nodes): batch_size = len(pos_samples) neg_batch_size = batch_size * self.k neg_samples = np.tile(pos_samples, (self.k, 1)) values = np.random.randint(num_nodes, size=neg_batch_size) choices = np.random.uniform(size=neg_batch_size) subj = choices > 0.5 obj = choices <= 0.5 neg_samples[subj, 0] = values[subj] neg_samples[obj, 2] = values[obj] samples = np.concatenate((pos_samples, neg_samples)) # binary labels indicating positive and negative samples labels = np.zeros(batch_size * (self.k + 1), dtype=np.float32) labels[:batch_size] = 1 return torch.from_numpy(samples), torch.from_numpy(labels) class SubgraphIterator: def __init__(self, g, num_rels, sample_size=30000, num_epochs=6000): self.g = g self.num_rels = num_rels self.sample_size = sample_size self.num_epochs = num_epochs self.pos_sampler = GlobalUniform(g, sample_size) self.neg_sampler = NegativeSampler() def __len__(self): return self.num_epochs def __getitem__(self, i): eids = self.pos_sampler.sample() src, dst = self.g.find_edges(eids) src, dst = src.numpy(), dst.numpy() rel = self.g.edata[dgl.ETYPE][eids].numpy() # relabel nodes to have consecutive node IDs uniq_v, edges = np.unique((src, dst), return_inverse=True) num_nodes = len(uniq_v) # edges is the concatenation of src, dst with relabeled ID src, dst = np.reshape(edges, (2, -1)) relabeled_data = np.stack((src, rel, dst)).transpose() samples, labels = self.neg_sampler.sample(relabeled_data, num_nodes) # use only half of the positive edges chosen_ids = np.random.choice(np.arange(self.sample_size), size=int(self.sample_size / 2), replace=False) src = src[chosen_ids] dst = dst[chosen_ids] rel = rel[chosen_ids] src, dst = np.concatenate((src, dst)), np.concatenate((dst, src)) rel = np.concatenate((rel, rel + self.num_rels)) sub_g = dgl.graph((src, dst), num_nodes=num_nodes) sub_g.edata[dgl.ETYPE] = torch.from_numpy(rel) sub_g.edata['norm'] = dgl.norm_by_dst(sub_g).unsqueeze(-1) uniq_v = torch.from_numpy(uniq_v).view(-1).long() return sub_g, uniq_v, samples, labels class RGCN(nn.Module): def __init__(self, num_nodes, h_dim, num_rels): super().__init__() # two-layer RGCN self.emb = nn.Embedding(num_nodes, h_dim) self.conv1 = RelGraphConv(h_dim, h_dim, num_rels, regularizer='bdd', num_bases=100, self_loop=True) self.conv2 = RelGraphConv(h_dim, h_dim, num_rels, regularizer='bdd', num_bases=100, self_loop=True) self.dropout = nn.Dropout(0.2) def forward(self, g, nids): x = self.emb(nids) h = F.relu(self.conv1(g, x, g.edata[dgl.ETYPE], g.edata['norm'])) h = self.dropout(h) h = self.conv2(g, h, g.edata[dgl.ETYPE], g.edata['norm']) return self.dropout(h) class LinkPredict(nn.Module): def __init__(self, num_nodes, num_rels, h_dim = 500, reg_param=0.01): super().__init__() self.rgcn = RGCN(num_nodes, h_dim, num_rels * 2) self.reg_param = reg_param self.w_relation = nn.Parameter(torch.Tensor(num_rels, h_dim)) nn.init.xavier_uniform_(self.w_relation, gain=nn.init.calculate_gain('relu')) def calc_score(self, embedding, triplets): s = embedding[triplets[:,0]] r = self.w_relation[triplets[:,1]] o = embedding[triplets[:,2]] score = torch.sum(s * r * o, dim=1) return score def forward(self, g, nids): return self.rgcn(g, nids) def regularization_loss(self, embedding): return torch.mean(embedding.pow(2)) + torch.mean(self.w_relation.pow(2)) def get_loss(self, embed, triplets, labels): # each row in the triplets is a 3-tuple of (source, relation, destination) score = self.calc_score(embed, triplets) predict_loss = F.binary_cross_entropy_with_logits(score, labels) reg_loss = self.regularization_loss(embed) return predict_loss + self.reg_param * reg_loss def filter(triplets_to_filter, target_s, target_r, target_o, num_nodes, filter_o=True): """Get candidate heads or tails to score""" target_s, target_r, target_o = int(target_s), int(target_r), int(target_o) # Add the ground truth node first if filter_o: candidate_nodes = [target_o] else: candidate_nodes = [target_s] for e in range(num_nodes): triplet = (target_s, target_r, e) if filter_o else (e, target_r, target_o) # Do not consider a node if it leads to a real triplet if triplet not in triplets_to_filter: candidate_nodes.append(e) return torch.LongTensor(candidate_nodes) def perturb_and_get_filtered_rank(emb, w, s, r, o, test_size, triplets_to_filter, filter_o=True): """Perturb subject or object in the triplets""" num_nodes = emb.shape[0] ranks = [] for idx in tqdm.tqdm(range(test_size), desc="Evaluate"): target_s = s[idx] target_r = r[idx] target_o = o[idx] candidate_nodes = filter(triplets_to_filter, target_s, target_r, target_o, num_nodes, filter_o=filter_o) if filter_o: emb_s = emb[target_s] emb_o = emb[candidate_nodes] else: emb_s = emb[candidate_nodes] emb_o = emb[target_o] target_idx = 0 emb_r = w[target_r] emb_triplet = emb_s * emb_r * emb_o scores = torch.sigmoid(torch.sum(emb_triplet, dim=1)) _, indices = torch.sort(scores, descending=True) rank = int((indices == target_idx).nonzero()) ranks.append(rank) return torch.LongTensor(ranks) def calc_mrr(emb, w, test_mask, triplets_to_filter, batch_size=100, filter=True): with torch.no_grad(): test_triplets = triplets_to_filter[test_mask] s, r, o = test_triplets[:,0], test_triplets[:,1], test_triplets[:,2] test_size = len(s) triplets_to_filter = {tuple(triplet) for triplet in triplets_to_filter.tolist()} ranks_s = perturb_and_get_filtered_rank(emb, w, s, r, o, test_size, triplets_to_filter, filter_o=False) ranks_o = perturb_and_get_filtered_rank(emb, w, s, r, o, test_size, triplets_to_filter) ranks = torch.cat([ranks_s, ranks_o]) ranks += 1 # change to 1-indexed mrr = torch.mean(1.0 / ranks.float()).item() mr = torch.mean(ranks.float()).item() print("MRR (filtered): {:.6f}".format(mrr)) print("MR (filtered): {:.6f}".format(mr)) hits=[1,3,10] for hit in hits: avg_count = torch.mean((ranks <= hit).float()) print("Hits (filtered) @ {}: {:.6f}".format(hit, avg_count.item())) return mrr def train(dataloader, test_g, test_nids, test_mask, triplets, device, model_state_file, model): optimizer = torch.optim.Adam(model.parameters(), lr=1e-2) best_mrr = 0 for epoch, batch_data in enumerate(dataloader): # single graph batch model.train() g, train_nids, edges, labels = batch_data g = g.to(device) train_nids = train_nids.to(device) edges = edges.to(device) labels = labels.to(device) embed = model(g, train_nids) loss = model.get_loss(embed, edges, labels) optimizer.zero_grad() loss.backward() nn.utils.clip_grad_norm_(model.parameters(), max_norm=1.0) # clip gradients optimizer.step() print("Epoch {:04d} | Loss {:.4f} | Best MRR {:.4f}".format(epoch, loss.item(), best_mrr)) if (epoch + 1) % 500 == 0: # perform validation on CPU because full graph is too large model = model.cpu() model.eval() embed = model(test_g, test_nids) mrr = calc_mrr(embed, model.w_relation, test_mask, triplets, batch_size=500) # save best model if best_mrr < mrr: best_mrr = mrr torch.save({'state_dict': model.state_dict(), 'epoch': epoch}, model_state_file) model = model.to(device) if __name__ == '__main__': device = torch.device('cuda' if torch.cuda.is_available() else 'cpu') print(f'Training with DGL built-in RGCN module') # load and preprocess dataset # data = FB15k237Dataset(reverse=False) data = MyDataset(raw_dir=r'data/FB15k237',reverse=False) g = data[0] num_nodes = g.num_nodes() num_rels = data.num_rels train_g = get_subset_g(g, g.edata['train_mask'], num_rels) test_g = get_subset_g(g, g.edata['train_mask'], num_rels, bidirected=True) test_g.edata['norm'] = dgl.norm_by_dst(test_g).unsqueeze(-1) test_nids = torch.arange(0, num_nodes) test_mask = g.edata['test_mask'] subg_iter = SubgraphIterator(train_g, num_rels) # uniform edge sampling dataloader = GraphDataLoader(subg_iter, batch_size=1, collate_fn=lambda x: x[0]) # Prepare data for metric computation src, dst = g.edges() triplets = torch.stack([src, g.edata['etype'], dst], dim=1) # create RGCN model model = LinkPredict(num_nodes, num_rels).to(device) # train model_state_file = 'model_state.pth' train(dataloader, test_g, test_nids, test_mask, triplets, device, model_state_file, model) # testing print("Testing...") checkpoint = torch.load(model_state_file) model = model.cpu() # test on CPU model.eval() model.load_state_dict(checkpoint['state_dict']) embed = model(test_g, test_nids) best_mrr = calc_mrr(embed, model.w_relation, test_mask, triplets, batch_size=500) print("Best MRR {:.4f} achieved using the epoch {:04d}".format(best_mrr, checkpoint['epoch']))- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
- 45
- 46
- 47
- 48
- 49
- 50
- 51
- 52
- 53
- 54
- 55
- 56
- 57
- 58
- 59
- 60
- 61
- 62
- 63
- 64
- 65
- 66
- 67
- 68
- 69
- 70
- 71
- 72
- 73
- 74
- 75
- 76
- 77
- 78
- 79
- 80
- 81
- 82
- 83
- 84
- 85
- 86
- 87
- 88
- 89
- 90
- 91
- 92
- 93
- 94
- 95
- 96
- 97
- 98
- 99
- 100
- 101
- 102
- 103
- 104
- 105
- 106
- 107
- 108
- 109
- 110
- 111
- 112
- 113
- 114
- 115
- 116
- 117
- 118
- 119
- 120
- 121
- 122
- 123
- 124
- 125
- 126
- 127
- 128
- 129
- 130
- 131
- 132
- 133
- 134
- 135
- 136
- 137
- 138
- 139
- 140
- 141
- 142
- 143
- 144
- 145
- 146
- 147
- 148
- 149
- 150
- 151
- 152
- 153
- 154
- 155
- 156
- 157
- 158
- 159
- 160
- 161
- 162
- 163
- 164
- 165
- 166
- 167
- 168
- 169
- 170
- 171
- 172
- 173
- 174
- 175
- 176
- 177
- 178
- 179
- 180
- 181
- 182
- 183
- 184
- 185
- 186
- 187
- 188
- 189
- 190
- 191
- 192
- 193
- 194
- 195
- 196
- 197
- 198
- 199
- 200
- 201
- 202
- 203
- 204
- 205
- 206
- 207
- 208
- 209
- 210
- 211
- 212
- 213
- 214
- 215
- 216
- 217
- 218
- 219
- 220
- 221
- 222
- 223
- 224
- 225
- 226
- 227
- 228
- 229
- 230
- 231
- 232
- 233
- 234
- 235
- 236
- 237
- 238
- 239
- 240
- 241
- 242
- 243
- 244
- 245
- 246
- 247
- 248
- 249
- 250
- 251
- 252
- 253
- 254
- 255
- 256
- 257
- 258
- 259
- 260
- 261
- 262
- 263
- 264
- 265
- 266
- 267
- 268
- 269
- 270
- 271
- 272
- 273
- 274
- 275
- 276
- 277
- 278
- 279
- 280
- 281
- 282
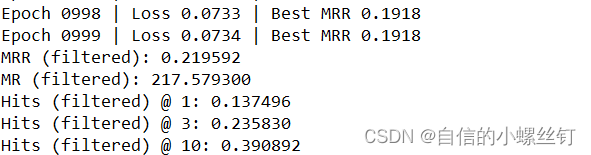
跑代码的输出图如下:
🆗,over!
-
相关阅读:
过程构成 中的小棱形 0..* 1..是什么意思过程构成 中的小棱形 0.. 1..*是什么意思
时空数据预测输入数据转换问题
mysql之行锁和表锁篇
【Linux系统编程:信号】产生信号 | 阻塞信号 | 处理信号 | 可重入函数
【C++】6-19 方阵的转置 分数 10
【智慧工地源码】基于AI视觉技术赋能智慧工地
西门子三菱等主流PLC如何通过4G无线网络来实现远程控制?
【SpringSecurity】十一、SpringSecurity集成JWT实现token的方法与校验
简析Acrel-1000安科瑞变电站综合自动化系统选型与应用
Java项目:SSM在线宿舍管理系统
- 原文地址:https://blog.csdn.net/qq_45791939/article/details/127975351
