-
【知识图谱】实践篇——基于医疗知识图谱的问答系统实践(Part2):图谱数据准备与导入
前序文章:
【知识图谱】实践篇——基于医疗知识图谱的问答系统实践(Part1):项目介绍与环境准备
背景
前文已经介绍了该系统的环境准备。下面介绍图谱数据获取,数据主要从:http://jib.xywy.com/ 爬取。
环境准备
按照原来的计划是将数据爬取相关的代码也过一下的,于是做了以下相关配置。
这里选择数据存储的库是mongodb。和以往一样,我依然使用docker容器化的方式去安装,相关方法可以参考:Docker 安装 MongoDB。然后安装连接mongodb的驱动程序:
pip install pymongo。因为需要连接数据库等,这里按照之前的做法,创建一个配置文件。
KGQAMedicine/data/config.ini[neo4j] host=http://192.168.56.101 port=7474 user=neo4j password=root [mongodb] host=http://192.168.56.101 port=27017 user=admin password=123456 [sys]- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
KGQAMedicine/utils/config.py
#!/usr/bin/python # -*- coding: UTF-8 -*- """ @author: juzipi @file: config.py @time:2022/07/20 @description: """ from configparser import ConfigParser class SysConfig(object): __doc__ = """ system config """ # 单例,全局唯一 def __new__(cls, *args, **kwargs): if not hasattr(SysConfig, '_instance'): SysConfig._instance = object.__new__(cls) return SysConfig._instance config_parser = ConfigParser() config_parser.read("./data/config.ini") # neo4j NEO4J_HOST = config_parser.get("neo4j", 'host') NEO4J_PORT = int(config_parser.get("neo4j", 'port')) NEO4J_USER = config_parser.get("neo4j", 'user') NEO4J_PASSWORD = config_parser.get('neo4j', 'password') # mongodb MONGODB_HOST = config_parser.get("mongodb", 'host') MONGODB_PORT = int(config_parser.get("mongodb", 'port')) MONGODB_USER = config_parser.get("mongodb", 'user') MONGODB_PASSWORD = config_parser.get('mongodb', 'password')- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
除此之外,我们还使用requests库以及lxml爬取数据以及解析页面。
但是考虑到由于原项目时间久远,原爬取的页面可能存在变化。这里介绍一下数据来源网站,如果有对相关数据感兴趣的话,可以在这个网址上爬取。我们使用原项目中已经爬取和处理完毕的数据。
数据来源
项目中数据来源于寻医问药网站,需要说明的是,网站也声明:不能作为诊断及医疗的依据。网站页面如下:

我将原项目中的数据放到KGQAMedicine/data/medicial.json中,并将路径配置的配置文件中。从数据形式上来看,该文件中的数据应该是从mongodb中导出。图谱数据导入
该部分改写内容代码如下:
KGQAMedicine/get_data/build_graph.py
import json import os import tqdm from py2neo import Graph, Node from utils.config import SysConfig class MedicalGraph(object): def __init__(self): self.data_path = SysConfig.DATA_ORIGIN_PATH self.graph = Graph(SysConfig.NEO4J_HOST + ":" + str(SysConfig.NEO4J_PORT), auth=(SysConfig.NEO4J_USER, SysConfig.NEO4J_PASSWORD)) self.raw_graph_data = None def _read_nodes(self): # 共7类节点 drugs = [] # 药品 foods = [] # 食物 checks = [] # 检查 departments = [] # 科室 producers = [] # 药品大类 diseases = [] # 疾病 symptoms = [] # 症状 disease_infos = [] # 疾病信息 # 构建节点实体关系 relation_department_department = [] # 科室-科室关系 relation_diseases_noteat = [] # 疾病-忌吃食物关系 relation_diseases_doeat = [] # 疾病-宜吃食物关系 relation_diseases_recommandeat = [] # 疾病-推荐吃食物关系 relation_diseases_commonddrug = [] # 疾病-通用药品关系 rels_recommanddrug = [] # 疾病-热门药品关系 rels_check = [] # 疾病-检查关系 relation_drug_producer = [] # 厂商-药物关系 rels_symptom = [] # 疾病症状关系 rels_acompany = [] # 疾病并发关系 rels_category = [] # 疾病与科室之间的关系 with open(self.data_path, 'r', encoding='utf8') as reader: for data in tqdm.tqdm(reader, desc=f"reading {self.data_path} fle"): disease_dict = {} data_json = json.loads(data) disease = data_json['name'] disease_dict['name'] = disease diseases.append(disease) disease_dict['desc'] = '' disease_dict['prevent'] = '' disease_dict['cause'] = '' disease_dict['easy_get'] = '' disease_dict['cure_department'] = '' disease_dict['cure_way'] = '' disease_dict['cure_lasttime'] = '' disease_dict['symptom'] = '' disease_dict['cured_prob'] = '' if 'symptom' in data_json: symptoms += data_json['symptom'] for symptom in data_json['symptom']: rels_symptom.append([disease, symptom]) if 'acompany' in data_json: for acompany in data_json['acompany']: rels_acompany.append([disease, acompany]) if 'desc' in data_json: disease_dict['desc'] = data_json['desc'] if 'prevent' in data_json: disease_dict['prevent'] = data_json['prevent'] if 'cause' in data_json: disease_dict['cause'] = data_json['cause'] if 'get_prob' in data_json: disease_dict['get_prob'] = data_json['get_prob'] if 'easy_get' in data_json: disease_dict['easy_get'] = data_json['easy_get'] if 'cure_department' in data_json: cure_department = data_json['cure_department'] if len(cure_department) == 1: rels_category.append([disease, cure_department[0]]) if len(cure_department) == 2: big = cure_department[0] small = cure_department[1] relation_department_department.append([small, big]) rels_category.append([disease, small]) disease_dict['cure_department'] = cure_department departments += cure_department if 'cure_way' in data_json: disease_dict['cure_way'] = data_json['cure_way'] if 'cure_lasttime' in data_json: disease_dict['cure_lasttime'] = data_json['cure_lasttime'] if 'cured_prob' in data_json: disease_dict['cured_prob'] = data_json['cured_prob'] if 'common_drug' in data_json: common_drug = data_json['common_drug'] for drug in common_drug: relation_diseases_commonddrug.append([disease, drug]) drugs += common_drug if 'recommand_drug' in data_json: recommand_drug = data_json['recommand_drug'] drugs += recommand_drug for drug in recommand_drug: rels_recommanddrug.append([disease, drug]) if 'not_eat' in data_json: not_eat = data_json['not_eat'] for _not in not_eat: relation_diseases_noteat.append([disease, _not]) foods += not_eat do_eat = data_json['do_eat'] for _do in do_eat: relation_diseases_doeat.append([disease, _do]) foods += do_eat recommand_eat = data_json['recommand_eat'] for _recommand in recommand_eat: relation_diseases_recommandeat.append([disease, _recommand]) foods += recommand_eat if 'check' in data_json: check = data_json['check'] for _check in check: rels_check.append([disease, _check]) checks += check if 'drug_detail' in data_json: drug_detail = data_json['drug_detail'] producer = [i.split('(')[0] for i in drug_detail] relation_drug_producer += [[i.split('(')[0], i.split('(')[-1].replace(')', '')] for i in drug_detail] producers += producer disease_infos.append(disease_dict) return set(drugs), set(foods), set(checks), set(departments), set(producers), set(symptoms), set(diseases), disease_infos, \ rels_check, relation_diseases_recommandeat, relation_diseases_noteat, relation_diseases_doeat, relation_department_department, relation_diseases_commonddrug, relation_drug_producer, rels_recommanddrug, \ rels_symptom, rels_acompany, rels_category def create_graph_nodes(self): if self.raw_graph_data is None: self.raw_graph_data = self._read_nodes() Drugs, Foods, Checks, Departments, Producers, Symptoms, Diseases, disease_infos = self.raw_graph_data[: 8] self.create_diseases_nodes(disease_infos) self.create_node('Drug', Drugs) self.create_node('Food', Foods) self.create_node('Check', Checks) self.create_node('Department', Departments) self.create_node('Producer', Producers) self.create_node('Symptom', Symptoms) def create_node(self, label, nodes): for node_name in tqdm.tqdm(nodes, desc=f"creating {label} nodes"): node = Node(label, name=node_name) self.graph.create(node) def create_diseases_nodes(self, disease_infos): """ 创建知识图谱中心疾病的节点 :param disease_infos: :return: """ for disease_dict in tqdm.tqdm(disease_infos, desc="creating diseases nodes"): node = Node("Disease", name=disease_dict['name'], desc=disease_dict['desc'], prevent=disease_dict['prevent'], cause=disease_dict['cause'], easy_get=disease_dict['easy_get'], cure_lasttime=disease_dict['cure_lasttime'], cure_department=disease_dict['cure_department'] , cure_way=disease_dict['cure_way'], cured_prob=disease_dict['cured_prob']) self.graph.create(node) def create_graph_relations(self): if self.raw_graph_data is None: self.raw_graph_data = self._read_nodes() rels_check, rels_recommandeat, rels_noteat, rels_doeat, rels_department, rels_commonddrug, rels_drug_producer, rels_recommanddrug, rels_symptom, rels_acompany, rels_category = self.raw_graph_data[ 8:] self.create_relationship('Disease', 'Food', rels_recommandeat, 'recommand_eat', '推荐食谱') self.create_relationship('Disease', 'Food', rels_noteat, 'no_eat', '忌吃') self.create_relationship('Disease', 'Food', rels_doeat, 'do_eat', '宜吃') self.create_relationship('Department', 'Department', rels_department, 'belongs_to', '属于') self.create_relationship('Disease', 'Drug', rels_commonddrug, 'common_drug', '常用药品') self.create_relationship('Producer', 'Drug', rels_drug_producer, 'drugs_of', '生产药品') self.create_relationship('Disease', 'Drug', rels_recommanddrug, 'recommand_drug', '好评药品') self.create_relationship('Disease', 'Check', rels_check, 'need_check', '诊断检查') self.create_relationship('Disease', 'Symptom', rels_symptom, 'has_symptom', '症状') self.create_relationship('Disease', 'Disease', rels_acompany, 'acompany_with', '并发症') self.create_relationship('Disease', 'Department', rels_category, 'belongs_to', '所属科室') def create_relationship(self, start_node, end_node, edges, rel_type, rel_name): """ 创建关系 :param start_node: :param end_node: :param edges: :param rel_type: :param rel_name: :return: """ # 去重处理 set_edges = [] for edge in edges: set_edges.append('###'.join(edge)) for edge in tqdm.tqdm(set(set_edges), desc=f"building edge {start_node} - {end_node} rel type {rel_type} rel name {rel_name}"): edge = edge.split('###') p = edge[0] q = edge[1] query = "match(p:%s),(q:%s) where p.name='%s'and q.name='%s' create (p)-[rel:%s{name:'%s'}]->(q)" % ( start_node, end_node, p, q, rel_type, rel_name) try: self.graph.run(query) except Exception as e: print(e) @staticmethod def _write(file_path, data_list): with open(file_path, 'w', encoding='utf8') as writer: writer.write("\n".join(data_list)) def export_data_dict(self): if self.raw_graph_data is None: self.raw_graph_data = self._read_nodes() Drugs, Foods, Checks, Departments, Producers, Symptoms, Diseases = self.raw_graph_data[: 7] self._write(os.path.join(SysConfig.DATA_DICT_DIR, "drug.txt"), list(Drugs)) self._write(os.path.join(SysConfig.DATA_DICT_DIR, "food.txt"), list(Foods)) self._write(os.path.join(SysConfig.DATA_DICT_DIR, "check.txt"), list(Checks)) self._write(os.path.join(SysConfig.DATA_DICT_DIR, "department.txt"), list(Departments)) self._write(os.path.join(SysConfig.DATA_DICT_DIR, "producer.txt"), list(Producers)) self._write(os.path.join(SysConfig.DATA_DICT_DIR, "symptom.txt"), list(Symptoms)) self._write(os.path.join(SysConfig.DATA_DICT_DIR, "disease.txt"), list(Diseases))- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
- 45
- 46
- 47
- 48
- 49
- 50
- 51
- 52
- 53
- 54
- 55
- 56
- 57
- 58
- 59
- 60
- 61
- 62
- 63
- 64
- 65
- 66
- 67
- 68
- 69
- 70
- 71
- 72
- 73
- 74
- 75
- 76
- 77
- 78
- 79
- 80
- 81
- 82
- 83
- 84
- 85
- 86
- 87
- 88
- 89
- 90
- 91
- 92
- 93
- 94
- 95
- 96
- 97
- 98
- 99
- 100
- 101
- 102
- 103
- 104
- 105
- 106
- 107
- 108
- 109
- 110
- 111
- 112
- 113
- 114
- 115
- 116
- 117
- 118
- 119
- 120
- 121
- 122
- 123
- 124
- 125
- 126
- 127
- 128
- 129
- 130
- 131
- 132
- 133
- 134
- 135
- 136
- 137
- 138
- 139
- 140
- 141
- 142
- 143
- 144
- 145
- 146
- 147
- 148
- 149
- 150
- 151
- 152
- 153
- 154
- 155
- 156
- 157
- 158
- 159
- 160
- 161
- 162
- 163
- 164
- 165
- 166
- 167
- 168
- 169
- 170
- 171
- 172
- 173
- 174
- 175
- 176
- 177
- 178
- 179
- 180
- 181
- 182
- 183
- 184
- 185
- 186
- 187
- 188
- 189
- 190
- 191
- 192
- 193
- 194
- 195
- 196
- 197
- 198
- 199
- 200
- 201
- 202
- 203
- 204
- 205
- 206
- 207
- 208
- 209
- 210
- 211
- 212
- 213
- 214
- 215
- 216
- 217
- 218
- 219
- 220
- 221
- 222
- 223
- 224
- 225
- 226
- 227
- 228
- 229
- 230
- 231
- 232
- 233
- 234
- 235
- 236
程序主要是改写了原程序。由于导入数据库时间比较长,这里就没有尝试运行导入到数据库模块程序,只将对应的实体输入到KGQAMedicine/data/dict目录下。有兴趣的朋友可以尝试运行导入。在第一篇文章导入数据的基础上,执行
MATCH p=()-->() RETURN p LIMIT 200查询图数据库结果如下: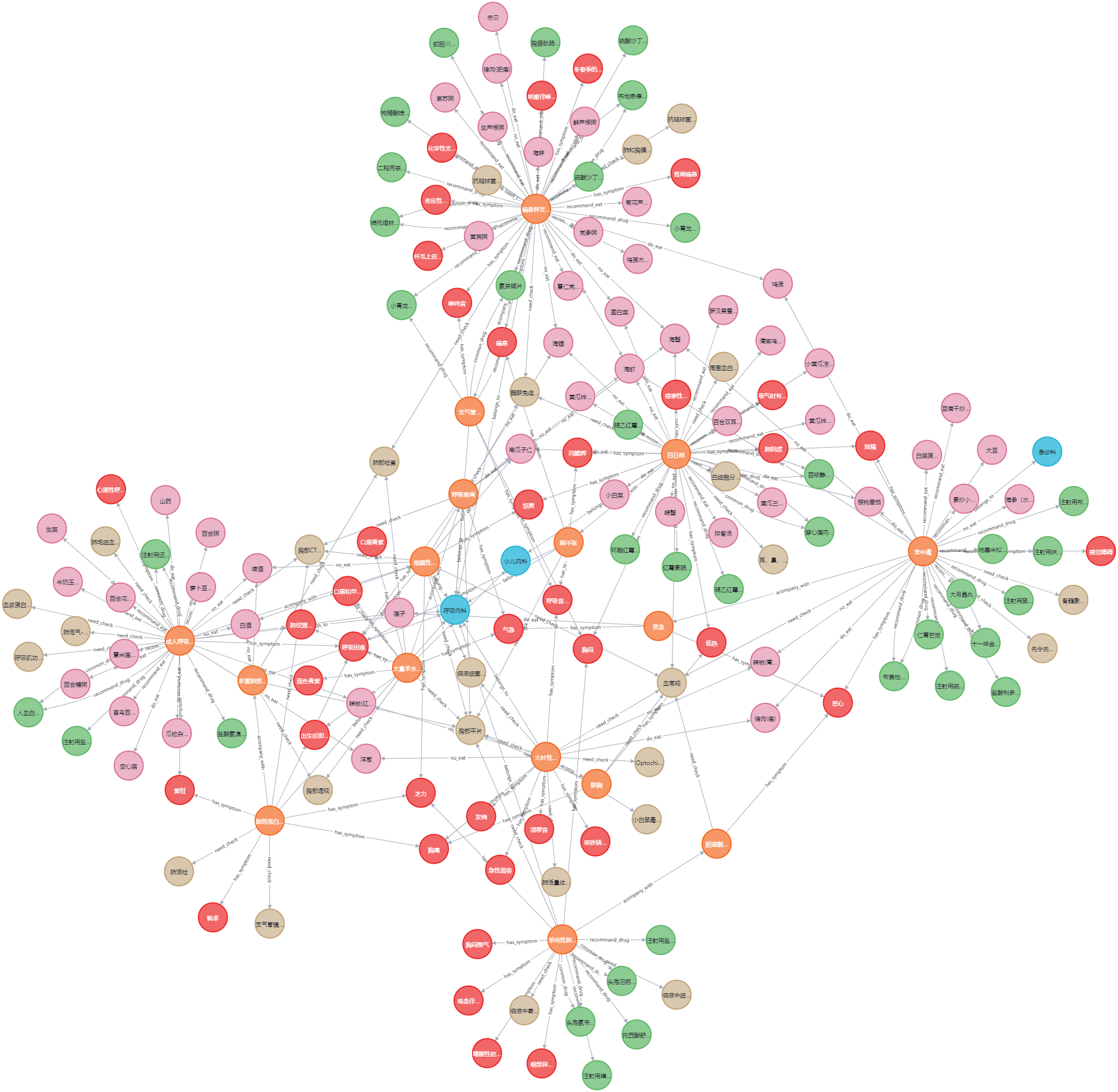
内容还是比较多的。 -
相关阅读:
求加django电影推荐系统中的推荐功能
Terra-Luna归零一年后:信任重建、加密未来路在何方?
Docker Cgroups资源控制
选择排序(Selection Sort)
渗透中 POC、EXP、Payload、Shellcode 的区别
【学习笔记】go协程和通道
opencv可以有多有趣
kaniko官方文档 - Build Images In Kubernetes
还在手撸TCP/UDP/COM通信?一个仅16K的库搞定!
Java动态代理
- 原文地址:https://blog.csdn.net/meiqi0538/article/details/125950332