-
【Coggle 30 Days of ML】糖尿病遗传风险检测挑战赛(2)
目录
3.K折交叉验证
任务
-
任务4:特征工程(使用pandas完成)
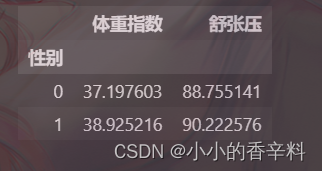
- 步骤1:统计每个性别对应的【体重指数】、【舒张压】平均值
- 步骤2:计算每个患者与每个性别平均值的差异;
- 步骤3:在上述基础上将训练集20%划分为验证集,使用逻辑回归完成训练,精度是否有提高?
- 步骤4:思考字段含义,尝试新的特征,将你的尝试写入博客;
-
任务5:特征筛选
- 步骤1:使用树模型完成模型的训练,通过特征重要性筛选出Top5的特征;
- 步骤2:使用筛选出的特征和逻辑回归进行训练,在验证集精度是否有提高?
- 步骤3:如果有提高,为什么?如果没有提高,为什么?
- 步骤4:将你的尝试写入博客;
-
任务7:多折训练与集成
- 步骤1:使用KFold完成数据划分;
- 步骤2:使用StratifiedKFold完成数据划分;
- 步骤3:使用StratifiedKFold配合LightGBM完成模型的训练和预测
- 步骤4:在步骤3训练得到了多少个模型,对测试集多次预测,将最新预测的结果文件提交到比赛,截图分数;
- 步骤5:使用交叉验证训练5个机器学习模型(svm、lr等),使用stacking完成集成,将最新预测的结果文件提交到比赛,截图分数;
Just Do It!
1.特征工程
特征工程部分很吃感觉,对于构建新的特征来说,试了好几个,构造出来效果反而更差了。emmm,只能说多尝试把。。。
- # 特征工程
- # 统计每个性别对应的【体重指数】、【舒张压】平均值
- # train_df.groupby('性别').describe()[['体重指数','舒张压']]
- train_df.groupby('性别').mean()[['体重指数','舒张压']]
- # train_df['体重指数-平均值']=train_df['体重指数']-train_df['性别'].map(
- # train_df.groupby('性别')['体重指数'].mean()
- # )
- # test_df['体重指数-平均值']=test_df['体重指数']-test_df['性别'].map(
- # test_df.groupby('性别')['体重指数'].mean()
- # )
- # test_df['体重指数-平均值']
- train_df['舒张压'].fillna(train_df['舒张压'].mean().round(2), inplace=True)
- test_df['舒张压'].fillna(train_df['舒张压'].mean().round(2), inplace=True)
- dict_糖尿病家族史 = {
- '无记录': 0,
- '叔叔或姑姑有一方患有糖尿病': 1,
- '叔叔或者姑姑有一方患有糖尿病': 1,
- '父母有一方患有糖尿病': 2
- }
- train_df['糖尿病家族史'] = train_df['糖尿病家族史'].map(dict_糖尿病家族史)
- test_df['糖尿病家族史'] = test_df['糖尿病家族史'].map(dict_糖尿病家族史)
- train_df['糖尿病家族史'] = train_df['糖尿病家族史'].astype('category')
- test_df['糖尿病家族史'] = train_df['糖尿病家族史'].astype('category')
- train_df['性别'] = train_df['性别'].astype('category')
- test_df['性别'] = train_df['性别'].astype('category')
- train_df['年龄'] = 2022 - train_df['出生年份']
- test_df['年龄'] = 2022 - test_df['出生年份']
- train_df['口服耐糖量测试_diff'] = train_df['口服耐糖量测试'] - train_df.groupby('糖尿病家族史').transform('mean')['口服耐糖量测试']
- test_df['口服耐糖量测试_diff'] = test_df['口服耐糖量测试'] - test_df.groupby('糖尿病家族史').transform('mean')['口服耐糖量测试']
- train_df['体重指数+肱三头肌皮褶厚度']=train_df['肱三头肌皮褶厚度']+train_df['体重指数']
- train_df
- test_df['体重指数+肱三头肌皮褶厚度']=test_df['肱三头肌皮褶厚度']+test_df['体重指数']
- test_df
接着可以进行标准化
- # 标准化
- train_df.loc[:,train_df.columns!='患有糖尿病标识'] = preprocessing.scale(train_df.drop('患有糖尿病标识',axis=1))
- train_df
为了方便后续模型融合,用到了xgboost,不支持类别型数据。因此需要将类别型的数据转换为数值型。
- train_df[['性别','糖尿病家族史']]=train_df[['性别','糖尿病家族史']].astype('int64')
- train_df.dtypes
- test_df[['性别','糖尿病家族史']]=test_df[['性别','糖尿病家族史']].astype('int64')
- test_df.dtypes

划分训练集和验证集
- # 把训练集划分为训练集和验证集,方便测试当前模型的好坏,并进行适当的调参
- train_df2,val_df,y_train,y_val = train_test_split(train_df.drop('患有糖尿病标识',axis=1),train_df['患有糖尿病标识'],test_size=0.2)
- train_df2
2.特征筛选
这部分其实通过树模型的feature_importances_属性就可以得到,然后再排个序,根据顺便挑选我们想要的前几个特征就可以了。
- # 特征筛选
- predictors = [i for i in train_df2.drop('编号',axis=1).columns]
- feat_imp = pd.Series(model.feature_importances_, predictors).sort_values(ascending=False)
- feat_imp
3.K折交叉验证
模型选择部分,用了lr、xgb、svm、lgbm做stacking模型融合。
- # 选择模型
- #
- # 使用逻辑回归进行训练
- model1 = make_pipeline(
- MinMaxScaler(),
- LogisticRegression()
- )
- # model.fit(train_df.drop('编号',axis=1),y_train)
- # 构建模型
- # model.fit(train_df.drop('编号',axis=1),y_train)
- # model3 = LGBMClassifier(
- # objective='binary',
- # max_depth=8,
- # device_type='gpu',
- # n_estimators=8000,
- # n_jobs=-1,
- # verbose=-1,
- # verbosity=-1,
- # learning_rate=0.1,
- # )
- # model.fit(train_df2.drop('编号',axis=1),y_train,eval_set=[(val_df.drop('编号',axis=1),y_val)])
- model2 = make_pipeline(
- MinMaxScaler(),
- XGBClassifier(
- n_estimators=4000,
- max_depth=5,
- random_state=42,
- device_type='gpu',
- early_stopping_rounds=30,
- n_jobs=-1,
- verbose=-1,
- verbosity=-1,
- )
- )
- model3 = make_pipeline(
- MinMaxScaler(),
- LGBMClassifier(
- objective='binary',
- max_depth=5,
- device_type='gpu',
- n_estimators=5000,
- n_jobs=-1,
- verbose=-1,
- verbosity=-1,
- learning_rate=0.1,
- )
- )
- model4 = make_pipeline(
- MinMaxScaler(),
- SVC(C=2,kernel='rbf',gamma=10,decision_function_shape='ovo')
- )
- estimators = [
- ('xgb', XGBClassifier(n_estimators=4000, random_state=42)),
- ('lr',model1),
- ('lgbm',model3),
- ('svm',model4)
- ]
- model = StackingClassifier(
- estimators=estimators
- )
K折交叉验证,此处K设置成了8。
- # K折交叉验证
- def k_fold(model, kf, X_tr, y, X_te, cate_col=None):
- train_pred = np.zeros( (len(X_tr), len(np.unique(y))) )
- test_pred = np.zeros( (len(X_te), len(np.unique(y))) )
- cv_clf = []
- for tr_idx, val_idx in kf.split(X_tr, y):
- x_tr = X_tr.iloc[tr_idx]; y_tr = y.iloc[tr_idx]
- x_val = X_tr.iloc[val_idx]; y_val = y.iloc[val_idx]
- eval_set = [(x_val, y_val)]
- model.fit(x_tr, y_tr)
- cv_clf.append(model)
- train_pred[val_idx] = model.predict_proba(x_val)
- test_pred += model.predict_proba(X_te)
- test_pred /= kf.n_splits
- return train_pred, test_pred, cv_clf
- train_pred, test_pred, cv_clf = k_fold(
- model, KFold(n_splits=8),
- train_df2.drop('编号',axis=1),
- y_train,
- val_df.drop('编号',axis=1),
- )
- val_df['label'] = test_pred.argmax(1)
分数反而低了。。。接着调参把。
OK,Fine!
-
-
相关阅读:
湖南湘潭家具3D轮廓扫描测量家居三维数字化外观逆向设计-CASAIM中科广电
Python学习第九篇:zipfile 库操作压缩包
Suc-Ala-Ala-Pro-Asp-pNA,CAS号: 165174-58-3
软件测试方法分类
c++day3
DetCLIP
SQLSERVER 查询正在执行的SQL语句
面向对象编程(二)
VS2022配置wxWidgets 3.0.5
信息化与信息系统5
- 原文地址:https://blog.csdn.net/doubleguy/article/details/125917466
