-
About Tolerance Intervals
It can be useful to have an upper and lower limit on data. These bounds can be used to help identify anomalies and set expectations for what to expect. A bound on observations from a population is called a tolerance interval.
A tolerance interval is different from a prediction interval that quantifies the uncertainty for a single predicted value. It is also different from a confidence interval that quantifies the uncertainty of a population parameter such as a mean. Instead, a tolerance interval covers a proportion of the population distribution.
you will know:
- That statistical tolerance intervals provide a bounds on observations from a population.
- That a tolerance interval requires that both a coverage proportion and confidence be specified.
- That the tolerance interval for a data sample with a Gaussian distribution can be easily calculated.
1.1 Tutorial Overview
1.Bounds on Data
2. What Are Statistical Tolerance Intervals?
3. How to Calculate Tolerance Intervals
4. Tolerance Interval for Gaussian Distribution
1.2 Bounds on Data
The range of common values for data is called a tolerance interval.
1.3 What Are Statistical Tolerance Intervals?
The tolerance interval is a bound on an estimate of the proportion of data in a population.
A statistical tolerance interval [contains] a specified proportion of the units from the sampled population or process.
A tolerance interval is defined in terms of two quantities:
- Coverage: The proportion of the population covered by the interval.
- Confidence: The probabilistic confidence that the interval covers the proportion of the population.
The tolerance interval is constructed from data using two coefficients, the coverage and the tolerance coefficient. The coverage is the proportion of the population (p) that the interval is supposed to contain. The tolerance coefficient is the degree of confidence with which the interval reaches the specified coverage.
1.4 How to Calculate Tolerance Intervals
The size of a tolerance interval is proportional to the size of the data sample from the population and the variance of the population. There are two main methods for calculating tolerance intervals depending on the distribution of data: parametric and nonparametric methods.
- Parametric Tolerance Interval: Use knowledge of the population distribution in specifying both the coverage and confidence. Often used to refer to a Gaussian distribution.
- Nonparametric Tolerance Interval: Use rank statistics to estimate the coverage and confidence, often resulting less precision (wider intervals) given the lack of information about the distribution.
Tolerance intervals are relatively straightforward to calculate for a sample of independent observations drawn from a Gaussian distribution. We will demonstrate this calculation in the next section.
1.5 Tolerance Interval for Gaussian Distribution
We will create a sample of 100 observations drawn from a Gaussian distribution with a mean of 50 and a standard deviation of 5.
- # generate dataset
- from numpy.random import randn
- data = 5 * randn(100) + 50
Remember that the degrees of freedom are the number of values in the calculation that can vary. Here, we have 100 observations, therefore 100 degrees of freedom. We do not know the standard deviation, therefore it must be estimated using the mean. This means our degrees of freedom will be (N - 1) or 99.
- # specify degrees of freedom
- n = len(data)
- dof = n - 1
Next, we must specify the proportional coverage of the data.
- # specify data coverage
- from scipy.stats import norm
- prop = 0.95
- prop_inv = (1.0 - prop) / 2.0
- gauss_critical = norm.ppf(prop_inv)
Next, we need to calculate the confidence of the coverage. We can do this by retrieving the critical value from the Chi-Squared distribution for the given number of degrees of freedom and desired probability. We can use the chi2.ppf() SciPy function.
- # specift confidence
- from scipy.stats import chi2
- prob = 0.99
- prop_inv = 1.0 - prob
- chi_critical = chi2.ppf(prop_inv,dof)
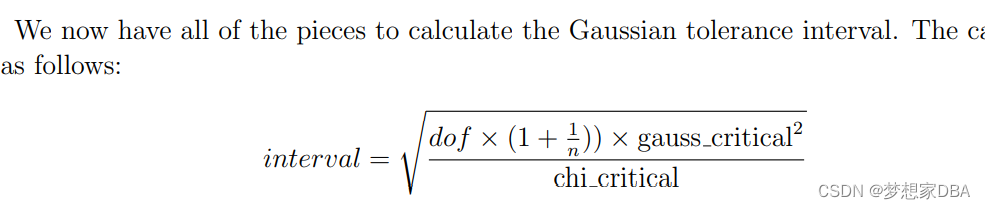
Where dof is the number of degrees of freedom, n is the size of the data sample, gauss critical is the critical value from the Gaussian distribution, such as 1.96 for 95% coverage of the population, and chi critical is the critical value from the Chi-Squared distribution for the desired confidence and degrees of freedom.
- # calculate tolerance interval
- from numpy import sqrt
- interval = sqrt((dof * (1 + (1/n)) * gauss_critical**2) / chi_critical)
We can tie all of this together and calculate the Gaussian tolerance interval for our data sample. The complete example is listed below.
- #parametric tolerance interval
- from numpy.random import seed
- from numpy.random import randn
- from numpy import mean
- from numpy import sqrt
- from scipy.stats import chi2
- from scipy.stats import norm
- # seed the random number generator
- seed(1)
- # generate dataset
- data = 5 * randn(100) + 50
- # specify degress of freedom
- n = len(data)
- dof = n - 1
- # specify data coverage
- prop = 0.95
- prop_inv = (1.0 - prop) / 2.0
- gauss_critical = norm.ppf(prop_inv)
- print('Gaussian critical value: %.3f (coverage=%d%%)' %(gauss_critical,prop*100))
- # specify confidence
- prob = 0.99
- prop_inv = 1.0 - prob
- chi_critical = chi2.ppf(prop_inv, dof)
- print('Chi-Squared critical value: %.3f (prob=%d%%,dof=%d)' %(chi_critical,prob*100,dof))
- # tolerance
- interval = sqrt((dof * (1 + (1/n)) * gauss_critical**2) / chi_critical)
- print('Tolerance Interval: %.3f' % interval)
- #summarize
- data_mean = mean(data)
- lower,upper = data_mean - interval, data_mean + interval
- print('%.2f to %.2f covers %d%% of data with a confidence of %d%%' %(lower,upper,prop*100,prob*100))
Running the example first calculates and prints the relevant critical values for the Gaussian and Chi-Squared distributions. The tolerance is printed, then presented correctly.

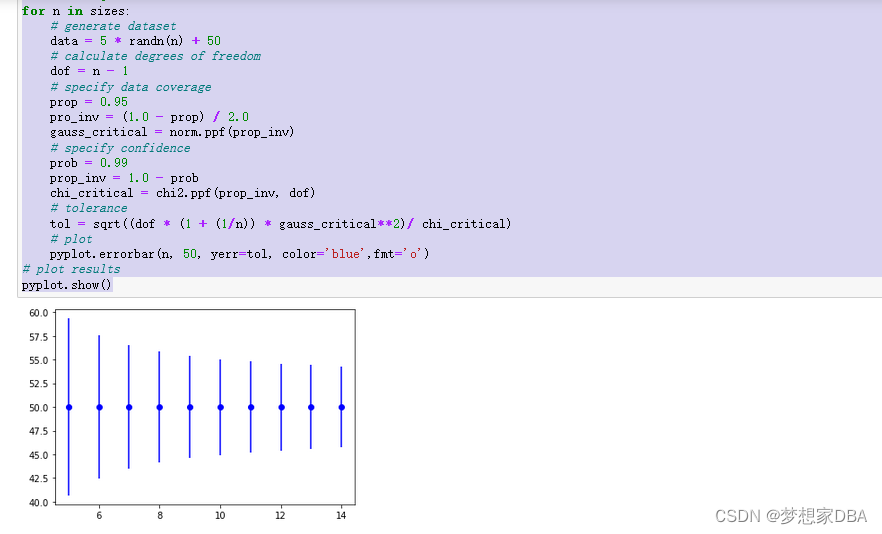
It can also be helpful to demonstrate how the tolerance interval will decrease (become more precise) as the size of the sample is increased. The example below demonstrates this by calculating the tolerance interval for different sample sizes for the same small contrived problem.
- # plot tolerance interval vs sample size
- from numpy.random import seed
- from numpy.random import randn
- from numpy import sqrt
- from scipy.stats import chi2
- from scipy.stats import norm
- from matplotlib import pyplot
- # seed the random number generator
- seed(1)
- # sample sizes
- seed(1)
- #sample sizes
- sizes = range(5,15)
- for n in sizes:
- # generate dataset
- data = 5 * randn(n) + 50
- # calculate degrees of freedom
- dof = n - 1
- # specify data coverage
- prop = 0.95
- pro_inv = (1.0 - prop) / 2.0
- gauss_critical = norm.ppf(prop_inv)
- # specify confidence
- prob = 0.99
- prop_inv = 1.0 - prob
- chi_critical = chi2.ppf(prop_inv, dof)
- # tolerance
- tol = sqrt((dof * (1 + (1/n)) * gauss_critical**2)/ chi_critical)
- # plot
- pyplot.errorbar(n, 50, yerr=tol, color='blue',fmt='o')
- # plot results
- pyplot.show()
Running the example creates a plot showing the tolerance interval around the true population mean. We can see that the interval becomes smaller (more precise) as the sample size is increased from 5 to 15 examples.
-
相关阅读:
[python]十九、进程、线程和协程
FL Studio 21.2.3.3586 for Mac中文版新功能介绍及2024年最新更新日志
C4D基础认识
WebAssembly学习笔记(一)
c++ 原子变量-Memory fence
真是学到了!——《Nginx的几个常用配置和技巧》
学习计划
【牛客网刷题系列 之 Verilog快速入门】~ 异步复位的串联T触发器、奇偶校验
哪些原因可能会导致 HBase 的 RegionServer 宕机?
[附源码]java毕业设计流浪动物领养系统
- 原文地址:https://blog.csdn.net/u011868279/article/details/125631615