-
批量从bam文件获取指定位置的碱基
批量从bam文件获取指定位置(RS.xlsx)的碱基后输出到一个excel,按照编号进行排序
- #!/user/bin/python3
- #coding:utf-8
- import pandas as pd
- import pysam
- import os,glob
- hetRatioCutoff=0.35
- primaryRatioCutoff=0.90
- baseRefL=['A', 'C', 'T', 'G']
- sampleGT={}
- def getGT(bam, chr_, pos_):
- bamfile = pysam.AlignmentFile(bam, "rb")
- depth={}
- totalDepth=0
- for pileupcolumn in bamfile.pileup(chr_, pos_-1, pos_+1):
- if pileupcolumn.reference_pos+1continueelif pileupcolumn.reference_pos+1>pos_:breakfor pileupread in pileupcolumn.pileups:if not pileupread.is_del and not pileupread.is_refskip:base=pileupread.alignment.query_sequence[pileupread.query_position].upper()depth[base]=1+depth[base] if base in depth else 1totalDepth+=1if totalDepth<=10: return 'NA' #低于10x时为No Calldepth=dict(sorted(depth.items(), key=lambda item:item[1], reverse=True))baseL=list(depth.keys())faL=[float(depth[i])/totalDepth for i in depth]if len(baseL)==1:return baseL[0]+baseL[0]else:if max(faL)>=primaryRatioCutoff:return baseL[faL.index(max(faL))]+baseL[faL.index(max(faL))]elif max(faL)>= hetRatioCutoff and max(faL) <=primaryRatioCutoff:faL_sort = sorted(faL,reverse=True)primary=faL_sort[0]+faL_sort[1]if primary>= primaryRatioCutoff:return baseL[faL.index(faL_sort[1])]+baseL[faL.index(faL_sort[0])] # 获取深度最大和第二大的碱基else:return 'NA'else:return 'NA'# read positiondf = pd.read_excel('RS.xlsx',header = 0,encoding = 'utf-8')df2 = []def output(bamname):barcode = os.path.splitext(bamname)[0]outfilename = barcode +'.xlsx'df1=df.to_dict(orient='records')for i in range(len(df1)):chr_ = df.iloc[i]['chr']pos_ = int(df.iloc[i]['pos'])rs = df.iloc[i]['rs']sampleGT[rs]=getGT(bamname, chr_, pos_)gt = 'gt_'+ barcodegenetype = 'genetype_'+ barcodedf1[i][gt] = sampleGT[rs]gt_list = list(df1[i][gt])if len(set(gt_list))== 1:if ''.join(set(gt_list)) == df.iloc[i]['ref']:df1[i][genetype]= "野生型"else:df1[i][genetype] = "纯合子"else:df1[i][genetype] = "杂合子"outDf=pd.DataFrame(df1)tempdf = outDf.loc[:,['rs',gt,genetype]]df2.append(tempdf)#outDf.to_excel(outfilename, index=False, columns=['rs','chr','pos','ref','alt',gt,genetype], encoding='utf-8')# 批量读取bam文件的前缀bamlist = glob.glob('*.bam')for bam in bamlist:output(bam)df3 = pd.concat(df2,axis=1, join='inner',sort = False) #concat合并可以是多个表作为一个list,merge合并是两个表df3 = df3.drop_duplicates().T.drop_duplicates().T # 去掉重复列'rs'outDf_result = pd.merge(df,df3,how='left',on='rs')outDf_result.to_excel('output.xlsx', index=False, encoding='utf-8')
以上办法是用pysam包处理,对于indel,质量不好等不一定准确,最好的还是用专业的软件处理,bcftools 和gatk。bacftools call snp后输出vcf文件后,再使用bacftools query -f命令处理vcf文件。
此处的test文件就是指定的位置,获取多个bam文件中这些位置的碱基
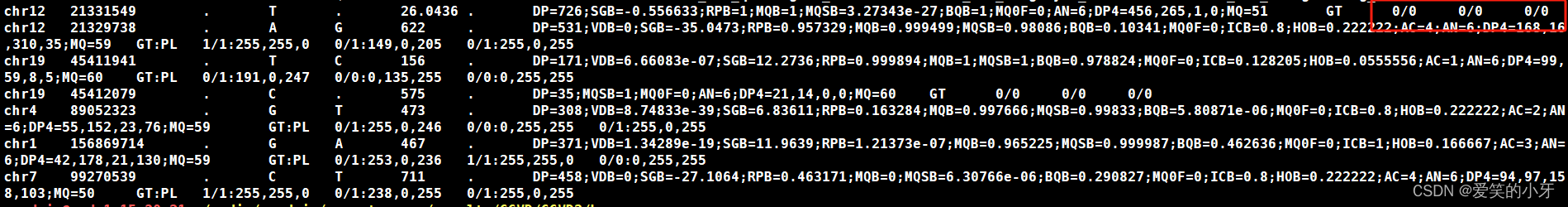
bcftools mpileup -R test -f /media/gsadmin/vd2/tmp/database/GATK/library/hg19/ucsc.hg19.fasta -q 10 -Q 10 --ff UNMAP -Ou *.bam | bcftools call -m >variants.vcfbcftools call -mv表示call为变异的位置碱基,如果不加v,就可以call出未变异的。
- 相关阅读:
Web前端:React对比Vue,哪一个最适合开发人员?
3000帧动画图解MySQL为什么需要binlog、redo log和undo log
Android 使用Camera2 API 和 GLSurfaceView实现相机预览
小程序面试题
问卷调查小程序功能清单
专项技能训练五《云计算网络技术与应用》实训6-2:ryu控制器安装
荷兰国旗问题与快速排序算法
kubernetes学习-概念3
什么是依赖注入(Dependency Injection)?它在 C++ 中是如何实现的?
C++ Qt 学习(五):Qt Web 编程
- 原文地址:https://blog.csdn.net/Cassiel60/article/details/133760509
