-
多输入多输出 | MATLAB实现GA-BP遗传算法优化BP神经网络多输入多输出
多输入多输出 | MATLAB实现GA-BP遗传算法优化BP神经网络多输入多输出
预测效果
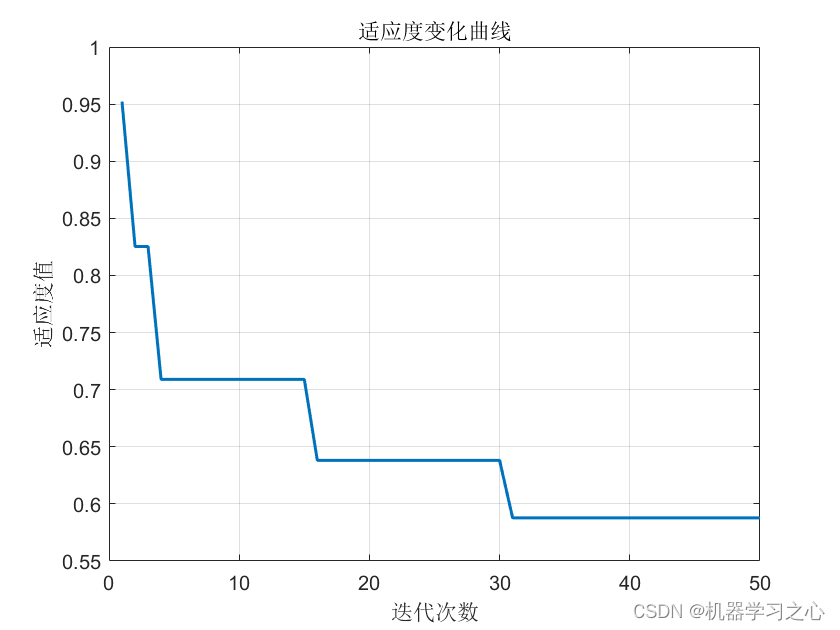
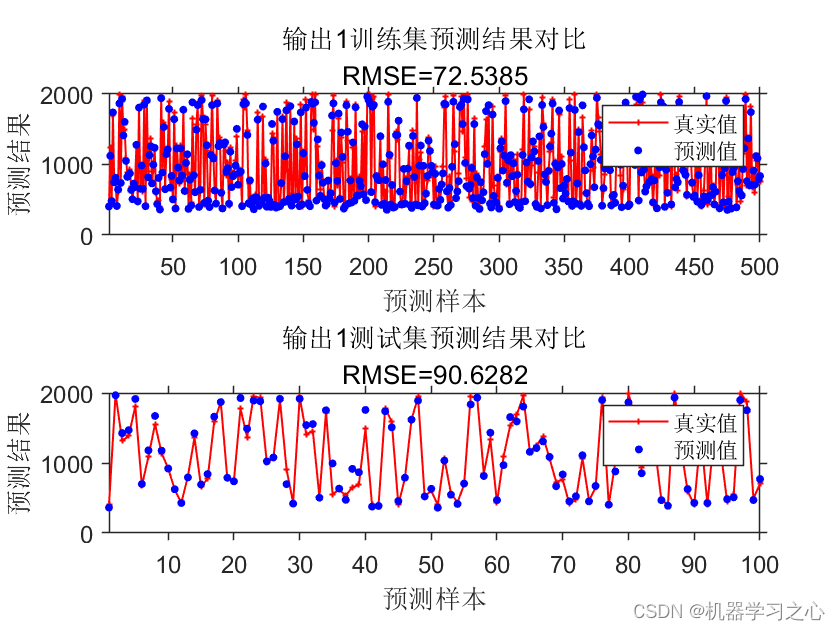
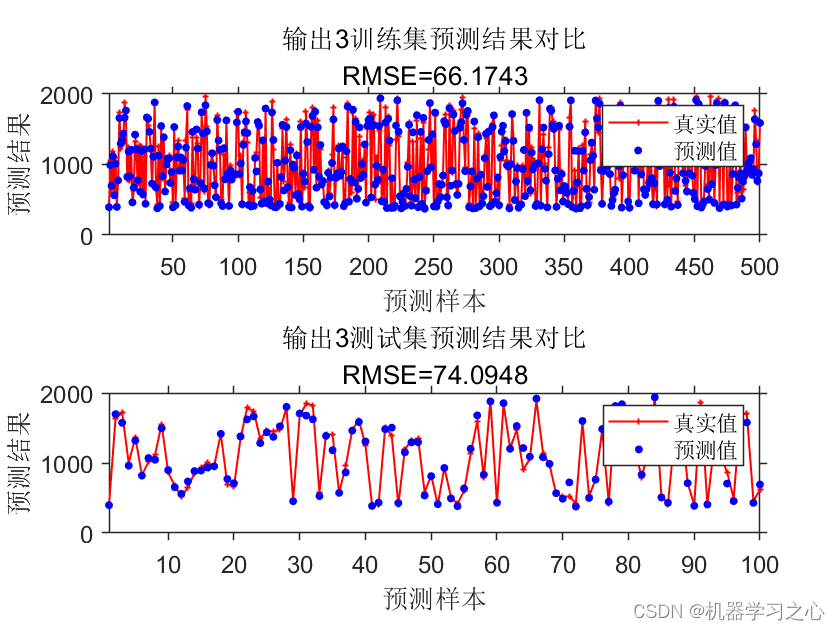


基本介绍
多输入多输出 | MATLAB实现GA-BP遗传算法优化BP神经网络多输入多输出
1.data为数据集,10个输入特征,3个输出变量。
2.main.m为主程序文件。
3.命令窗口输出MBE、MAE和R2,可在下载区获取数据和程序内容。程序设计
- 完整程序和数据下载方式:私信博主回复MATLAB实现GA-BP遗传算法优化BP神经网络多输入多输出。
%----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- function [x, endPop, bPop, traceInfo] = ga(bounds, evalFN, evalOps, startPop, opts, ... termFN, termOps, selectFN, selectOps, xOverFNs, xOverOps, mutFNs, mutOps) % Output Arguments: % x - the best solution found during the course of the run % endPop - the final population % bPop - a trace of the best population % traceInfo - a matrix of best and means of the ga for each generation % % Input Arguments: % bounds - a matrix of upper and lower bounds on the variables % evalFN - the name of the evaluation .m function % evalOps - options to pass to the evaluation function ([NULL]) % startPop - a matrix of solutions that can be initialized % from initialize.m % opts - [epsilon prob_ops display] change required to consider two % solutions different, prob_ops 0 if you want to apply the % genetic operators probabilisticly to each solution, 1 if % you are supplying a deterministic number of operator % applications and display is 1 to output progress 0 for % quiet. ([1e-6 1 0]) % termFN - name of the .m termination function (['maxGenTerm']) % termOps - options string to be passed to the termination function % ([100]). % selectFN - name of the .m selection function (['normGeomSelect']) % selectOpts - options string to be passed to select after % select(pop,#,opts) ([0.08]) % xOverFNS - a string containing blank seperated names of Xover.m % files (['arithXover heuristicXover simpleXover']) % xOverOps - A matrix of options to pass to Xover.m files with the % first column being the number of that xOver to perform % similiarly for mutation ([2 0;2 3;2 0]) % mutFNs - a string containing blank seperated names of mutation.m % files (['boundaryMutation multiNonUnifMutation ... % nonUnifMutation unifMutation']) % mutOps - A matrix of options to pass to Xover.m files with the % first column being the number of that xOver to perform % similiarly for mutation ([4 0 0;6 100 3;4 100 3;4 0 0]) %% 初始化参数 n = nargin; if n < 2 || n == 6 || n == 10 || n == 12 disp('Insufficient arguements') end % 默认评估选项 if n < 3 evalOps = []; end % 默认参数 if n < 5 opts = [1e-6, 1, 0]; end % 默认参数 if isempty(opts) opts = [1e-6, 1, 0]; end %% 判断是否为m文件 if any(evalFN < 48) % 浮点数编码 if opts(2) == 1 e1str = ['x=c1; c1(xZomeLength)=', evalFN ';']; e2str = ['x=c2; c2(xZomeLength)=', evalFN ';']; % 二进制编码 else e1str = ['x=b2f(endPop(j,:),bounds,bits); endPop(j,xZomeLength)=', evalFN ';']; end else % 浮点数编码 if opts(2) == 1 e1str = ['[c1 c1(xZomeLength)]=' evalFN '(c1,[gen evalOps]);']; e2str = ['[c2 c2(xZomeLength)]=' evalFN '(c2,[gen evalOps]);']; % 二进制编码 else e1str=['x=b2f(endPop(j,:),bounds,bits);[x v]=' evalFN ... '(x,[gen evalOps]); endPop(j,:)=[f2b(x,bounds,bits) v];']; end end %% 默认终止信息 if n < 6 termOps = 100; termFN = 'maxGenTerm'; end %% 默认变异信息 if n < 12 % 浮点数编码 if opts(2) == 1 mutFNs = 'boundaryMutation multiNonUnifMutation nonUnifMutation unifMutation'; mutOps = [4, 0, 0; 6, termOps(1), 3; 4, termOps(1), 3;4, 0, 0]; % 二进制编码 else mutFNs = 'binaryMutation'; mutOps = 0.05; end end %% 默认交叉信息 if n < 10 % 浮点数编码 if opts(2) == 1 xOverFNs = 'arithXover heuristicXover simpleXover'; xOverOps = [2, 0; 2, 3; 2, 0]; % 二进制编码 else xOverFNs = 'simpleXover'; xOverOps = 0.6; end end %% 仅默认选择选项,即轮盘赌。 if n < 9 selectOps = []; end %% 默认选择信息 if n < 8 selectFN = 'normGeomSelect'; selectOps = 0.08; end %% 默认终止信息 if n < 6 termOps = 100; termFN = 'maxGenTerm'; end %% 没有定的初始种群 if n < 4 startPop = []; end %% 随机生成种群 if isempty(startPop) startPop = initializega(80, bounds, evalFN, evalOps, opts(1: 2)); end %% 二进制编码 if opts(2) == 0 bits = calcbits(bounds, opts(1)); end %% 参数设置 xOverFNs = parse(xOverFNs); mutFNs = parse(mutFNs); xZomeLength = size(startPop, 2); % xzome 的长度 numVar = xZomeLength - 1; % 变量数 popSize = size(startPop,1); % 种群人口个数 endPop = zeros(popSize, xZomeLength); % 第二种群矩阵 numXOvers = size(xOverFNs, 1); % Number of Crossover operators numMuts = size(mutFNs, 1); % Number of Mutation operators epsilon = opts(1); % Threshold for two fittness to differ oval = max(startPop(:, xZomeLength)); % Best value in start pop bFoundIn = 1; % Number of times best has changed done = 0; % Done with simulated evolution gen = 1; % Current Generation Number collectTrace = (nargout > 3); % Should we collect info every gen floatGA = opts(2) == 1; % Probabilistic application of ops display = opts(3); % Display progress %% 精英模型 while(~done) [bval, bindx] = max(startPop(:, xZomeLength)); % Best of current pop best = startPop(bindx, :); if collectTrace traceInfo(gen, 1) = gen; % current generation traceInfo(gen, 2) = startPop(bindx, xZomeLength); % Best fittness traceInfo(gen, 3) = mean(startPop(:, xZomeLength)); % Avg fittness traceInfo(gen, 4) = std(startPop(:, xZomeLength)); end %% 最佳解 if ( (abs(bval - oval) > epsilon) || (gen==1)) % 更新显示 if display fprintf(1, '\n%d %f\n', gen, bval); end % 更新种群矩阵 if floatGA bPop(bFoundIn, :) = [gen, startPop(bindx, :)]; else bPop(bFoundIn, :) = [gen, b2f(startPop(bindx, 1 : numVar), bounds, bits)... startPop(bindx, xZomeLength)]; end bFoundIn = bFoundIn + 1; % Update number of changes oval = bval; % Update the best val else if display fprintf(1,'%d ',gen); % Otherwise just update num gen end end %% 选择种群 endPop = feval(selectFN, startPop, [gen, selectOps]); % 以参数为操作数的模型运行 if floatGA for i = 1 : numXOvers for j = 1 : xOverOps(i, 1) a = round(rand * (popSize - 1) + 1); % Pick a parent b = round(rand * (popSize - 1) + 1); % Pick another parent xN = deblank(xOverFNs(i, :)); % Get the name of crossover function [c1, c2] = feval(xN, endPop(a, :), endPop(b, :), bounds, [gen, xOverOps(i, :)]); % Make sure we created a new if c1(1 : numVar) == endPop(a, (1 : numVar)) c1(xZomeLength) = endPop(a, xZomeLength); elseif c1(1:numVar) == endPop(b, (1 : numVar)) c1(xZomeLength) = endPop(b, xZomeLength); else eval(e1str); end if c2(1 : numVar) == endPop(a, (1 : numVar)) c2(xZomeLength) = endPop(a, xZomeLength); elseif c2(1 : numVar) == endPop(b, (1 : numVar)) c2(xZomeLength) = endPop(b, xZomeLength); else eval(e2str); end endPop(a, :) = c1; endPop(b, :) = c2; end end for i = 1 : numMuts for j = 1 : mutOps(i, 1) a = round(rand * (popSize - 1) + 1); c1 = feval(deblank(mutFNs(i, :)), endPop(a, :), bounds, [gen, mutOps(i, :)]); if c1(1 : numVar) == endPop(a, (1 : numVar)) c1(xZomeLength) = endPop(a, xZomeLength); else eval(e1str); end endPop(a, :) = c1; end end %% 运行遗传算子的概率模型 else for i = 1 : numXOvers xN = deblank(xOverFNs(i, :)); cp = find((rand(popSize, 1) < xOverOps(i, 1)) == 1); if rem(size(cp, 1), 2) cp = cp(1 : (size(cp, 1) - 1)); end cp = reshape(cp, size(cp, 1) / 2, 2); for j = 1 : size(cp, 1) a = cp(j, 1); b = cp(j, 2); [endPop(a, :), endPop(b, :)] = feval(xN, endPop(a, :), endPop(b, :), ... bounds, [gen, xOverOps(i, :)]); end end for i = 1 : numMuts mN = deblank(mutFNs(i, :)); for j = 1 : popSize endPop(j, :) = feval(mN, endPop(j, :), bounds, [gen, mutOps(i, :)]); eval(e1str); end end end % 更新记录 gen = gen + 1; done = feval(termFN, [gen, termOps], bPop, endPop); % See if the ga is done startPop = endPop; % Swap the populations [~, bindx] = min(startPop(:, xZomeLength)); % Keep the best solution startPop(bindx, :) = best; % replace it with the worst end [bval, bindx] = max(startPop(:, xZomeLength)); %% 显示结果 if display fprintf(1, '\n%d %f\n', gen, bval); end %% 二进制编码 x = startPop(bindx, :); if opts(2) == 0 x = b2f(x, bounds,bits); bPop(bFoundIn, :) = [gen, b2f(startPop(bindx, 1 : numVar), bounds, bits)... startPop(bindx, xZomeLength)]; else bPop(bFoundIn, :) = [gen, startPop(bindx, :)]; end %% 赋值 if collectTrace traceInfo(gen, 1) = gen; % 当前迭代次数 traceInfo(gen, 2) = startPop(bindx, xZomeLength); % 最佳适应度 traceInfo(gen, 3) = mean(startPop(:, xZomeLength)); % 平均适应度 end- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
- 45
- 46
- 47
- 48
- 49
- 50
- 51
- 52
- 53
- 54
- 55
- 56
- 57
- 58
- 59
- 60
- 61
- 62
- 63
- 64
- 65
- 66
- 67
- 68
- 69
- 70
- 71
- 72
- 73
- 74
- 75
- 76
- 77
- 78
- 79
- 80
- 81
- 82
- 83
- 84
- 85
- 86
- 87
- 88
- 89
- 90
- 91
- 92
- 93
- 94
- 95
- 96
- 97
- 98
- 99
- 100
- 101
- 102
- 103
- 104
- 105
- 106
- 107
- 108
- 109
- 110
- 111
- 112
- 113
- 114
- 115
- 116
- 117
- 118
- 119
- 120
- 121
- 122
- 123
- 124
- 125
- 126
- 127
- 128
- 129
- 130
- 131
- 132
- 133
- 134
- 135
- 136
- 137
- 138
- 139
- 140
- 141
- 142
- 143
- 144
- 145
- 146
- 147
- 148
- 149
- 150
- 151
- 152
- 153
- 154
- 155
- 156
- 157
- 158
- 159
- 160
- 161
- 162
- 163
- 164
- 165
- 166
- 167
- 168
- 169
- 170
- 171
- 172
- 173
- 174
- 175
- 176
- 177
- 178
- 179
- 180
- 181
- 182
- 183
- 184
- 185
- 186
- 187
- 188
- 189
- 190
- 191
- 192
- 193
- 194
- 195
- 196
- 197
- 198
- 199
- 200
- 201
- 202
- 203
- 204
- 205
- 206
- 207
- 208
- 209
- 210
- 211
- 212
- 213
- 214
- 215
- 216
- 217
- 218
- 219
- 220
- 221
- 222
- 223
- 224
- 225
- 226
- 227
- 228
- 229
- 230
- 231
- 232
- 233
- 234
- 235
- 236
- 237
- 238
- 239
- 240
- 241
- 242
- 243
- 244
- 245
- 246
- 247
- 248
- 249
- 250
- 251
- 252
- 253
- 254
- 255
- 256
- 257
- 258
- 259
- 260
- 261
- 262
- 263
- 264
- 265
- 266
- 267
- 268
- 269
- 270
- 271
- 272
- 273
- 274
- 275
- 276
- 277
- 278
- 279
- 280
- 281
- 282
- 283
- 284
- 285
- 286
- 287
- 288
- 289
- 290
- 291
- 292
- 293
- 294
- 295
- 296
- 297
- 298
- 299
- 300
- 301
- 302
- 303
- 304
- 305
- 306
- 307
往期精彩
MATLAB实现RBF径向基神经网络多输入多输出预测
MATLAB实现BP神经网络多输入多输出预测
MATLAB实现DNN神经网络多输入多输出预测参考资料
[1] https://blog.csdn.net/kjm13182345320/article/details/116377961
[2] https://blog.csdn.net/kjm13182345320/article/details/127931217
[3] https://blog.csdn.net/kjm13182345320/article/details/127894261 -
相关阅读:
串行原理编程,中文编程工具中的串行构件,串行连接操作简单
Linq to SQL语句之Top/Bottom和Paging分页和SqlMethods
每日晨会,或1-3-5晨会(项目开发进度把控)
EN 13859-2防水用柔性薄板—CE认证
2023最新SSM计算机毕业设计选题大全(附源码+LW)之java创新实践学分管理系统08a30
09_SpingBoot 集成Dubbo
设计模式——责任链模式(Chain of Responsibility Pattern)+ Spring相关源码
vmtouch——Linux下的文件缓存管理神器
各种测试方法,黑盒测试、白盒测试,静态测试,动态测试
程序员开发过程中还是需要写注释的
- 原文地址:https://blog.csdn.net/kjm13182345320/article/details/132957756
