-
Topic 20. 临床预测模型之竞争风险模型及计算生存概率
点击关注,桓峰基因
桓峰基因公众号推出基于R语言临床预测模型教程并配有视频在线教程,目前整理出来的教程目录如下:
Topic 10. 单因素 Logistic 回归分析—单因素分析表格
Topic 12 临床预测模型—列线表 (Nomogram)
Topic 13. 临床预测模型—一致性指数 (C-index)
Topic 14. 临床预测模型之校准曲线 (Calibration curve)
Topic 16. 临床预测模型之接收者操作特征曲线 (ROC)
Topic 19. 临床预测模型之输出每个患者列线图得分 (nomogramFormula)
上期介绍了Cox回归模型计算数据集中单个患者的列线图的得分、线性预测值。Cox回归模型的数学公式是我们计算的依据。接下来介绍如何计算生存概率?
前 言
竞争风险模型( Competing Risk Model ) : 指的是在观察队列中,存在某种已知事件可能会影响另一种事件发生的概率或者是完全阻碍其发生,则可认为前者与后者存在竞争风险。
什么叫竞争风险事件:
竞争风险事件:研究中结局事件可能有多个,某些结局将阻止感兴趣事件的出现或影响其发生的概率,各结局事件形成"竞争”关系,互为竞争风险事件。
例如慢性肾病患者死亡与透析、心肌梗死患者导致的死亡与其他死因, 生殖细胞癌患者死亡与继发恶性肿瘤,先天性心脏病患者术后死亡 与随访终点肺静脉梗阻存在竞争风险。
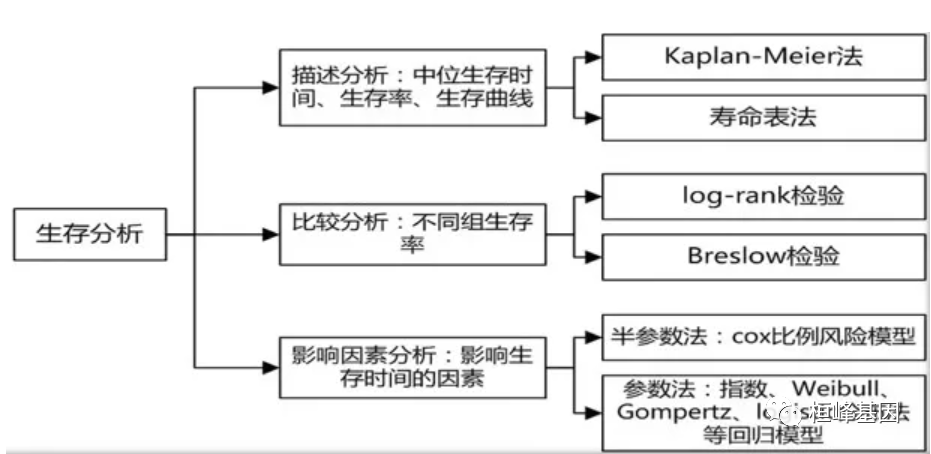
传统生存分析有哪些?
1、Kaplan-Meier法 ,适用于分组生存资料的分析,估计生存概率,需要知道患者的生存时间与状态
2、寿命表法:适用于未分组的生存资料,样本量较大,不需要知道患者的生存时间与状态;
3、Log-rank检验 :适用于两组或多组生存资料的比较;
4、cox回归:用于肿瘤和其他慢性病的预后因素分析。
为什么选用竞争风险模型?
传统的生存分析( survival analysis ) 一般只关心一个终点事件(即研究者感兴趣的结局)。将发生复发前死亡的个体、失访个体和未发生复发的个体均按删失数据( Censored Data)处理,要求个体删失情况与个体终点事件相互独立,结局不存在竞争风险。
竞争风险模型:适用于多个终点的生存数据,是一种处理多种潜在结局生存数据的分析方法,通过计算每个结局的累积发生率函数( Cumulative Incidences Function , CIF )进行分析。
单一结局的生存分析,主要关注研究对象的生存概率或者死L_风险,这里的生存概率仅仅是这一个结局的生存的概率,而竞争风险中,可能是多个结局共同的总的生存概率或者总的死L_风险,研究的兴趣点的生存概率是这个总生存概率的一部分。
在竞争风险模型中预测事件概率(累积发生率)是必要的。
软件包安装
我们这期主要使用 pec软件包和riskRegression软件包,安装如下:
if(!require(pec)) install.packages("pec") if(!require(riskRegression)) install.packages("riskRegression")- 1
- 2
- 3
- 4
- 5
参数说明
预测生存概率
从各种建模方法中提取生存概率预测。最突出的是考克斯回归模型,它可以用“coxph”和“cph”来拟合。
函数从各种建模方法中提取事件概率预测。其中最突出的是可以用pec包中的函数predictSurvProb()预测生存概率。
object: 一个拟合的模型,从中提取预测的生存概率;
newdata: 包含预测变量组合的数据框,用于计算预测的生存概率;
times: 响应变量范围内的时间向量,例如,响应是生存对象的时间,在此时间返回生存概率。
例子实操
例子一
Cox回归
函数自带的例子, 使用 prodlim 软件包生成生存数据:
library(rms) library(pec) # generate some survival data library(prodlim) set.seed(100) d <- SimSurv(100)- 1
- 2
- 3
- 4
- 5
- 6
- 7
构建Cox回归模型并计算预测生存概率:
# then fit a Cox model library(rms) coxmodel <- cph(Surv(time, status) ~ X1 + X2, data = d, surv = TRUE) coxmodel ## Cox Proportional Hazards Model ## ## cph(formula = Surv(time, status) ~ X1 + X2, data = d, surv = TRUE) ## ## Model Tests Discrimination ## Indexes ## Obs 100 LR chi2 41.58 R2 0.344 ## Events 58 d.f. 2 R2(2,100)0.327 ## Center 0.4893 Pr(> chi2) 0.0000 R2(2,58) 0.495 ## Score chi2 39.66 Dxy 0.465 ## Pr(> chi2) 0.0000 ## ## Coef S.E. Wald Z Pr(>|Z|) ## X1 0.9541 0.2839 3.36 0.0008 ## X2 0.9321 0.1653 5.64 <0.0001 ## # Extract predicted survival probabilities at selected time-points: ttt <- quantile(d$time) # for selected predictor values: ndat <- data.frame(X1 = c(0.25, 0.25, -0.05, 0.05), X2 = c(0, 1, 0, 1)) # as follows predictSurvProb(coxmodel, newdata = ndat, times = ttt) ## 0.4260983 2.1691480 4.9274490 7.6507514 14.4062674 ## 1 0.9955287 0.8986157 0.7441616 0.4961380 0.0160256016 ## 2 0.9886826 0.7622278 0.4721214 0.1686068 0.0000275735 ## 3 0.9966398 0.9228473 0.8009592 0.5907036 0.0448385378 ## 4 0.9906394 0.7990404 0.5378687 0.2297102 0.0001708277- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
分层构建Cox生存回归模型并计算预测生存概率
# stratified cox model sfit <- coxph(Surv(time, status) ~ strata(X1) + X2, data = d, , x = TRUE, y = TRUE) predictSurvProb(sfit, newdata = d[1:3, ], times = c(1, 3, 5, 10)) ## [,1] [,2] [,3] [,4] ## [1,] 0.9841264 0.9150092 0.8655247 0.5451199 ## [2,] 0.9360099 0.6617560 0.4824460 0.0845529 ## [3,] 0.9468067 0.7108774 0.5474556 0.1297774- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
再次模拟一套数据作为训练集和验证集,构建模型后计算预测生存概率,五个时间点的概率。获得一个有存活概率的矩阵,一列代表5个时间点,一行代表每个验证集个体:
## simulate some learning and some validation data learndat <- SimSurv(100) valdat <- SimSurv(100) ## use the learning data to fit a Cox model library(survival) fitCox <- coxph(Surv(time, status) ~ X1 + X2, data = learndat, x = TRUE, y = TRUE) fitCox ## Call: ## coxph(formula = Surv(time, status) ~ X1 + X2, data = learndat, ## x = TRUE, y = TRUE) ## ## coef exp(coef) se(coef) z p ## X1 0.9019 2.4642 0.2591 3.480 0.000501 ## X2 0.8777 2.4054 0.1663 5.278 1.31e-07 ## ## Likelihood ratio test=38.55 on 2 df, p=4.247e-09 ## n= 100, number of events= 65 ## suppose we want to predict the survival probabilities for all patients in ## the validation data at the following time points: 0, 12, 24, 36, 48, 60 psurv <- predictSurvProb(fitCox, newdata = valdat, times = seq(0, 60, 12)) head(psurv) ## [,1] [,2] [,3] [,4] [,5] [,6] ## [1,] 1 0.314747935 NA NA NA NA ## [2,] 1 0.004391524 NA NA NA NA ## [3,] 1 0.290617260 NA NA NA NA ## [4,] 1 0.736042412 NA NA NA NA ## [5,] 1 0.126687345 NA NA NA NA ## [6,] 1 0.296940557 NA NA NA NA ## This is a matrix with survival probabilities one column for each of the 5 ## time points one row for each validation set individual- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
当在coxph或survreg模型公式中使用时,指定脊回归项。可能性被减去/2乘以系数平方和。如果scale=T,则根据预测因子的缩放来计算系数的惩罚,使其具有单位方差。如果指定了df,则根据近似的自由度选择。
脊回归
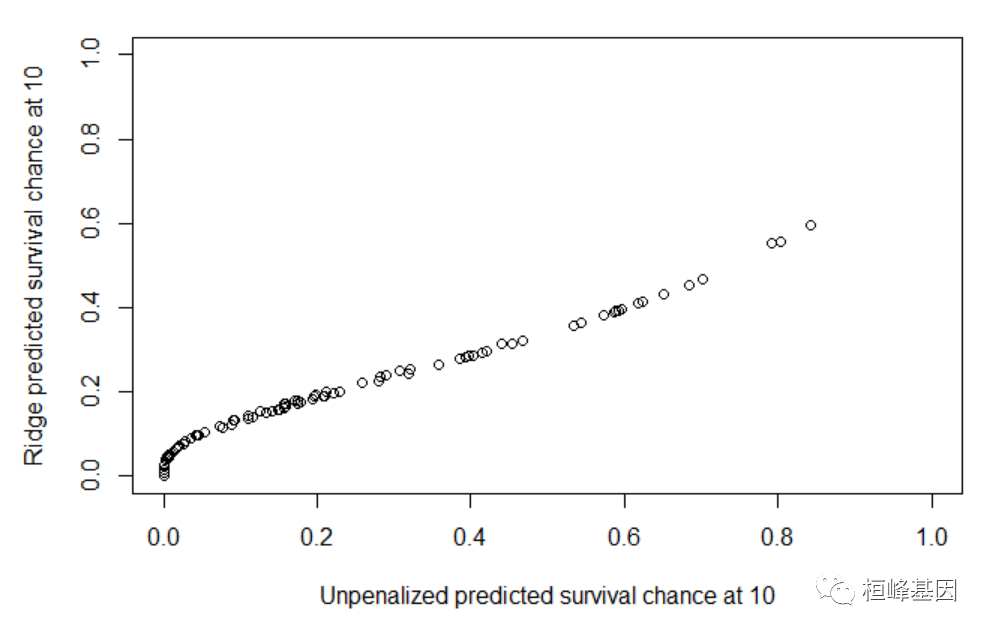
## Cox with ridge option f1 <- coxph(Surv(time, status) ~ X1 + X2, data = learndat, x = TRUE, y = TRUE) f2 <- coxph(Surv(time, status) ~ ridge(X1) + ridge(X2), data = learndat, x = TRUE, y = TRUE) plot(predictSurvProb(f1, newdata = valdat, times = 10), pec:::predictSurvProb.coxph(f2, newdata = valdat, times = 10), xlim = c(0, 1), ylim = c(0, 1), xlab = "Unpenalized predicted survival chance at 10", ylab = "Ridge predicted survival chance at 10")- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
例子二
我们仍然采用lung数据集,对数据进行分割为测试集和验证集:
library(sampling) lung = na.omit(lung) str(lung) ## 'data.frame': 167 obs. of 10 variables: ## $ inst : num 3 5 12 7 11 1 7 6 12 22 ... ## $ time : num 455 210 1022 310 361 ... ## $ status : num 2 2 1 2 2 2 2 2 2 2 ... ## $ age : num 68 57 74 68 71 53 61 57 57 70 ... ## $ sex : num 1 1 1 2 2 1 1 1 1 1 ... ## $ ph.ecog : num 0 1 1 2 2 1 2 1 1 1 ... ## $ ph.karno : num 90 90 50 70 60 70 70 80 80 90 ... ## $ pat.karno: num 90 60 80 60 80 80 70 80 70 100 ... ## $ meal.cal : num 1225 1150 513 384 538 ... ## $ wt.loss : num 15 11 0 10 1 16 34 27 60 -5 ... ## - attr(*, "na.action")= 'omit' Named int [1:61] 1 3 5 12 13 14 16 20 23 25 ... ## ..- attr(*, "names")= chr [1:61] "1" "3" "5" "12" ... set.seed(123) # 每层抽取70%的数据 train_id <- strata(lung, "status", size = rev(round(table(lung$status) * 0.7)))$ID_unit # 训练数据 train_data <- lung[train_id, ] # 测试数据 test_data <- lung[-train_id, ] # 查看训练、测试数据中正负样本比例 prop.table(table(train_data$status)) ## ## 1 2 ## 0.2820513 0.7179487 prop.table(table(test_data$status)) ## ## 1 2 ## 0.28 0.72- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
下面我们继续使用pec包中predictSurvProb()函数计算训练集、验证集与全集中各患者的365天的生存概率,并做为新的列加入到对应的数据集中,代码如下:
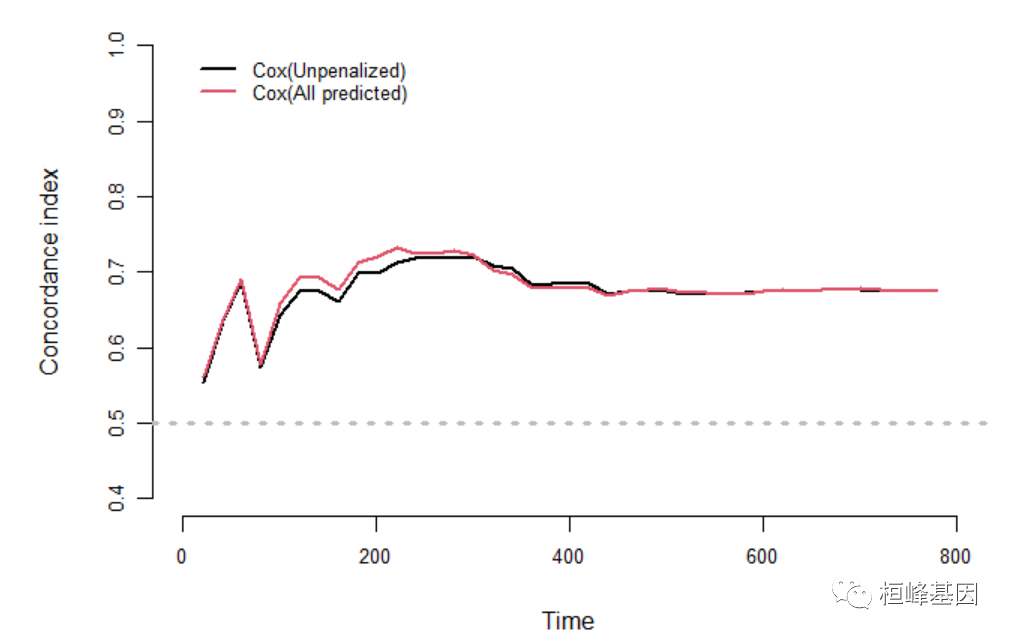
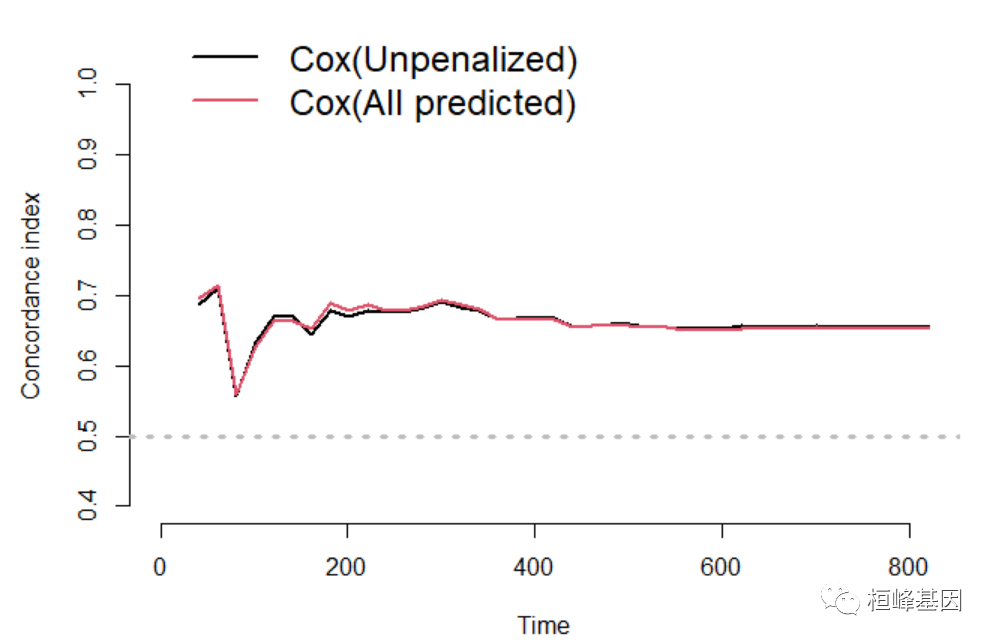
dd <- datadist(train_data) options(datadist = "dd") library(survival) f1 <- coxph(Surv(time, status) ~ age + sex + ph.ecog + ph.karno, data = train_data, x = TRUE, y = TRUE) f2 <- coxph(Surv(time, status) ~ ridge(age) + ridge(sex) + ridge(ph.ecog) + ridge(ph.karno), data = train_data, x = TRUE, y = TRUE) f3 <- coxph(Surv(time, status) ~ ., data = train_data, x = TRUE, y = TRUE) t = seq(1, 1000, 20) c_index <- pec::cindex(list(`Cox(Unpenalized)` = f1, `Cox(All predicted)` = f3), formula = Surv(time, status == 2) ~ ., data = train_data, eval.times = t) par(mgp = c(3.1, 0.8, 0), mar = c(5, 5, 3, 1), cex.axis = 0.8, cex.main = 0.8) plot(c_index, xlim = c(0, 800), legend.x = 1, legend.y = 1, legend.cex = 0.8)- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
下面我们继续使用pec包中predictSurvProb()函数计算训练集、验证集与全集中各患者的不同时间点的生存概率,并做为新的列加入到对应的数据集中,代码如下:
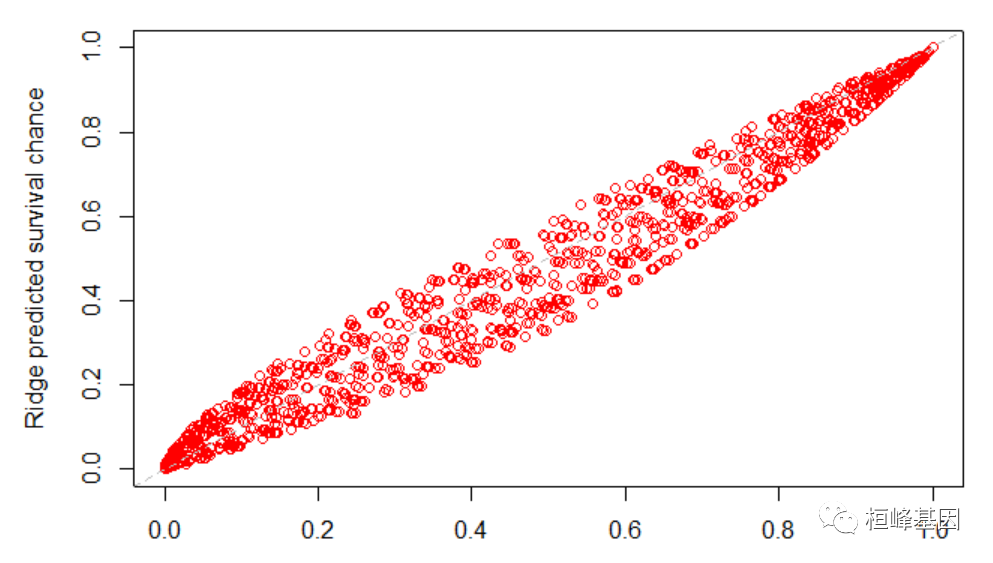
library(data.table) test_data = as.data.table(test_data) t = seq(1, max(train_data$time), 20) plot(predictSurvProb(f1, newdata = test_data, times = t), pec:::predictSurvProb.coxph(f2, newdata = test_data, times = t), xlim = c(0, 1), ylim = c(0, 1), col = "red", xlab = "Unpenalized predicted survival chance", ylab = "Ridge predicted survival chance") abline(a = 0, b = 1, lty = 2, col = "gray")- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
计算 C-index 在不同时间点的 bootstrap-crossvalidation 估计
bcvCindex <- pec::cindex(list(`Cox(Unpenalized)` = f1, `Cox(All predicted)` = f3), formula = Surv(time, status == 2) ~ ., data = train_data, splitMethod = "bootcv", B = 5, eval.times = t) print(bcvCindex) ## ## The c-index for right censored event times ## ## Prediction models: ## ## Cox(Unpenalized) Cox(All predicted) ## Cox(Unpenalized) Cox(All predicted) ## ## Right-censored response of a survival model ## ## No.Observations: 117 ## ## Pattern: ## Freq ## event 84 ## right.censored 33 ## ## Censoring model for IPCW: marginal model (Kaplan-Meier for censoring distribution) ## ## Method for estimating the prediction error: ## ## Bootstrap cross-validation ## ## Type: resampling ## Bootstrap sample size: 117 ## No. bootstrap samples: 5 ## Sample size: 117 ## ## Estimated C-index in % ## ## $AppCindex ## time=1 time=21 time=41 time=61 time=81 time=101 time=121 ## Cox(Unpenalized) NaN 55.3 63.4 68.7 57.4 64.2 67.5 ## Cox(All predicted) NaN 56.1 63.7 69.0 57.7 65.9 69.2 ## time=141 time=161 time=181 time=201 time=221 time=241 ## Cox(Unpenalized) 67.5 66.0 69.9 69.9 71.3 71.8 ## Cox(All predicted) 69.2 67.6 71.3 72.0 73.3 72.5 ## time=261 time=281 time=301 time=321 time=341 time=361 ## Cox(Unpenalized) 71.8 71.9 72.1 70.8 70.5 68.4 ## Cox(All predicted) 72.5 72.8 72.2 70.2 69.6 68.0 ## time=381 time=401 time=421 time=441 time=461 time=481 ## Cox(Unpenalized) 68.4 68.4 68.4 67.1 67.4 67.6 ## Cox(All predicted) 67.9 67.9 67.9 66.9 67.5 67.7 ## time=501 time=521 time=541 time=561 time=581 time=601 ## Cox(Unpenalized) 67.6 67.1 67.1 67.1 67.2 67.4 ## Cox(All predicted) 67.7 67.3 67.3 67.1 67.1 67.4 ## time=621 time=641 time=661 time=681 time=701 time=721 ## Cox(Unpenalized) 67.6 67.5 67.6 67.6 67.8 67.6 ## Cox(All predicted) 67.6 67.5 67.7 67.7 67.8 67.6 ## time=741 time=761 time=781 time=801 time=821 ## Cox(Unpenalized) 67.6 67.6 67.5 67.6 67.6 ## Cox(All predicted) 67.6 67.6 67.5 67.6 67.6 ## ## $BootCvCindex ## time=1 time=21 time=41 time=61 time=81 time=101 time=121 ## Cox(Unpenalized) NaN NaN 68.8 71.0 55.6 63.3 66.9 ## Cox(All predicted) NaN NaN 69.6 71.4 55.8 62.6 66.4 ## time=141 time=161 time=181 time=201 time=221 time=241 ## Cox(Unpenalized) 66.9 64.5 67.9 67.0 67.9 67.7 ## Cox(All predicted) 66.4 65.2 68.8 67.9 68.7 67.8 ## time=261 time=281 time=301 time=321 time=341 time=361 ## Cox(Unpenalized) 67.7 68.2 69.1 68.3 67.8 66.5 ## Cox(All predicted) 67.8 68.5 69.3 68.6 68.1 66.5 ## time=381 time=401 time=421 time=441 time=461 time=481 ## Cox(Unpenalized) 66.7 66.7 66.7 65.5 65.7 65.8 ## Cox(All predicted) 66.6 66.6 66.6 65.4 65.6 65.7 ## time=501 time=521 time=541 time=561 time=581 time=601 ## Cox(Unpenalized) 65.8 65.6 65.6 65.4 65.2 65.4 ## Cox(All predicted) 65.7 65.4 65.4 65.1 65.0 65.1 ## time=621 time=641 time=661 time=681 time=701 time=721 ## Cox(Unpenalized) 65.6 65.5 65.6 65.6 65.6 65.5 ## Cox(All predicted) 65.4 65.3 65.3 65.3 65.4 65.3 ## time=741 time=761 time=781 time=801 time=821 ## Cox(Unpenalized) 65.5 65.5 65.5 65.5 65.5 ## Cox(All predicted) 65.3 65.3 65.2 65.2 65.2 ## ## ## AppCindex : Apparent (training data) performance ## BootCvCindex : Bootstrap crossvalidated performance plot(bcvCindex)- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
- 45
- 46
- 47
- 48
- 49
- 50
- 51
- 52
- 53
- 54
- 55
- 56
- 57
- 58
- 59
- 60
- 61
- 62
- 63
- 64
- 65
- 66
- 67
- 68
- 69
- 70
- 71
- 72
- 73
- 74
- 75
- 76
- 77
- 78
- 79
- 80
- 81
- 82
- 83
- 84
- 85
- 86
竞争风险
对风险指标和风险预测模型的预测性能进行评分的方法
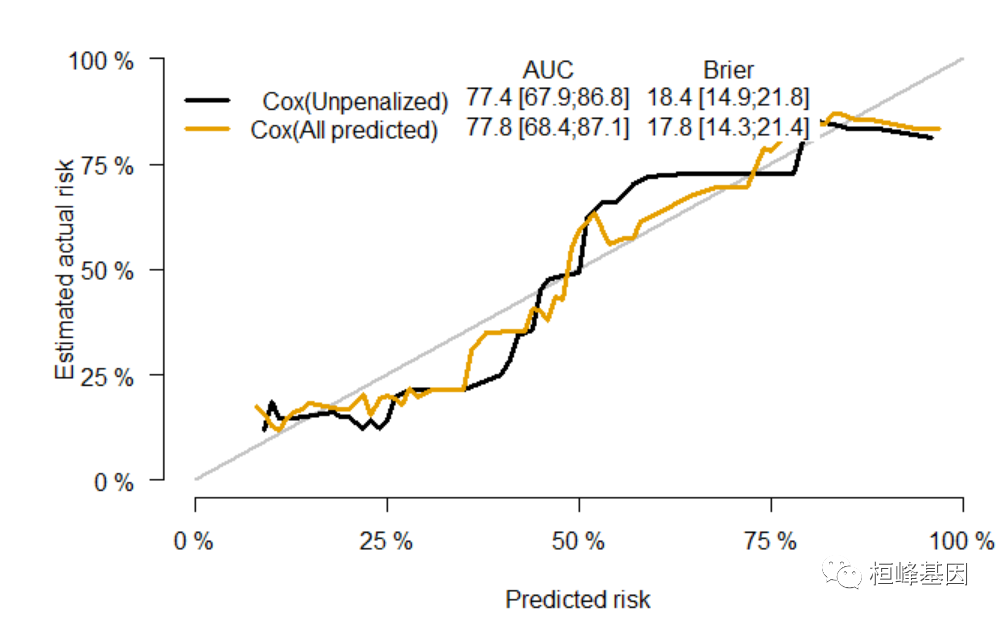
绘制 calibration curve
library(riskRegression) xb = Score(list(`Cox(Unpenalized)` = f1, `Cox(All predicted)` = f3), formula = Surv(time, status == 2) ~ age + sex + ph.ecog + ph.karno, data = train_data, plots = "calibration") plotCalibration(xb)- 1
- 2
- 3
- 4
- 5
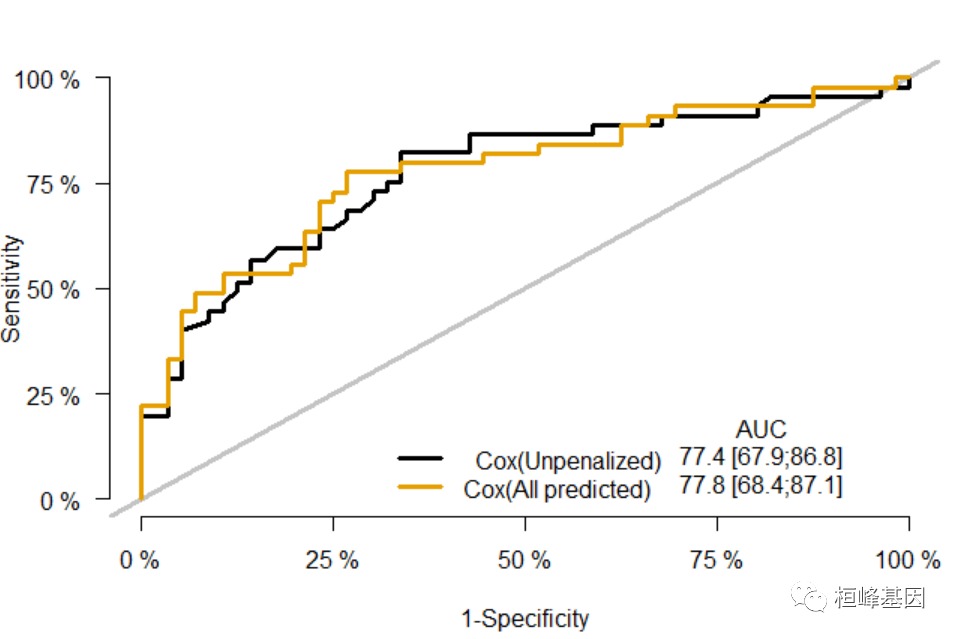
绘制 ROC 曲线
xb = Score(list(`Cox(Unpenalized)` = f1, `Cox(All predicted)` = f3), formula = Surv(time, status == 2) ~ age + sex + ph.ecog + ph.karno, data = train_data, plots = "ROC") plotROC(xb)- 1
- 2
- 3
- 4
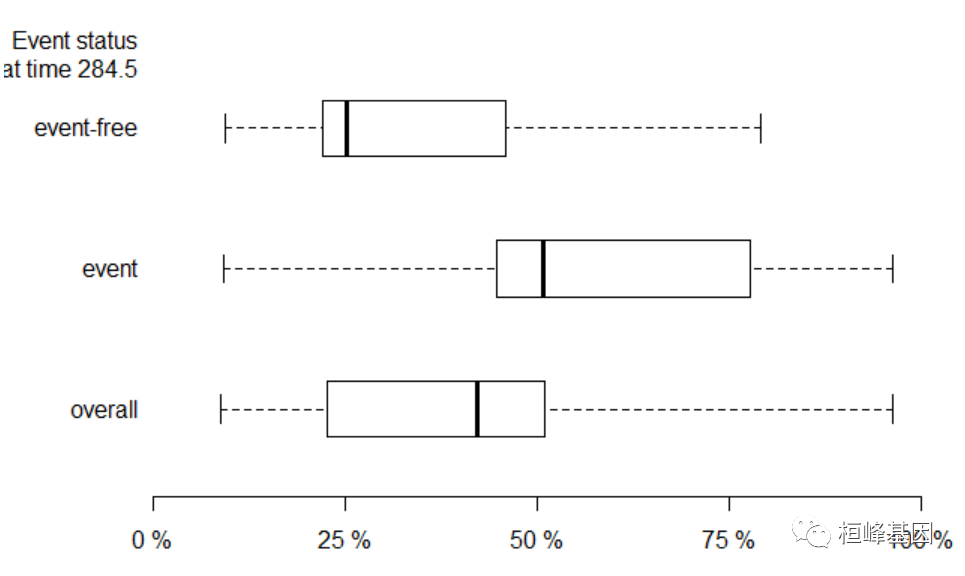
绘制箱线图
xb = Score(list(`Cox(Unpenalized)` = f1, `Cox(All predicted)` = f3), formula = Surv(time, status == 2) ~ age + sex + ph.ecog + ph.karno, data = train_data, plots = "boxplot") boxplot(xb)- 1
- 2
- 3
- 4
训练数据中的交叉验证模型
xb = Score(list(`Cox(Unpenalized)` = f1, `Cox(All predicted)` = f3), formula = Surv(time, status == 2) ~ age + sex + ph.ecog + ph.karno, data = train_data, split.method = "loob", B = 100, plots = "calibration") plotCalibration(xb)- 1
- 2
- 3
- 4
- 5
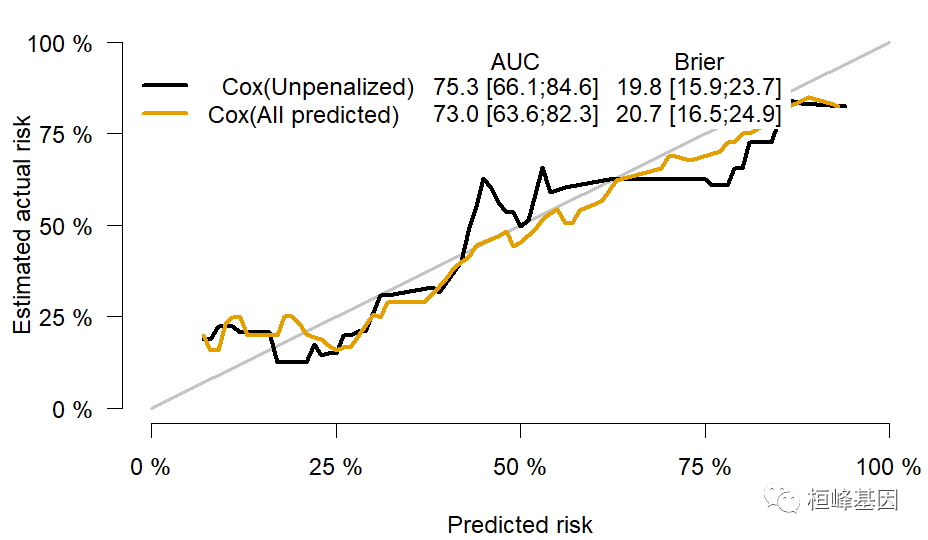
对于未经截尾的数据,其结果与从 Hmisc 中 rcorr.cens 函数得到的结果相同。
绘制预测风险
# survival library(survival) xs = Score(list(`Cox(Unpenalized)` = f1, `Cox(All predicted)` = f3), formula = Surv(time, status == 2) ~ 1, data = train_data, summary = "risks", null.model = 0L, times = c(365, 2 * 365)) plotRisk(xs, times = c(365, 2 * 365))- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
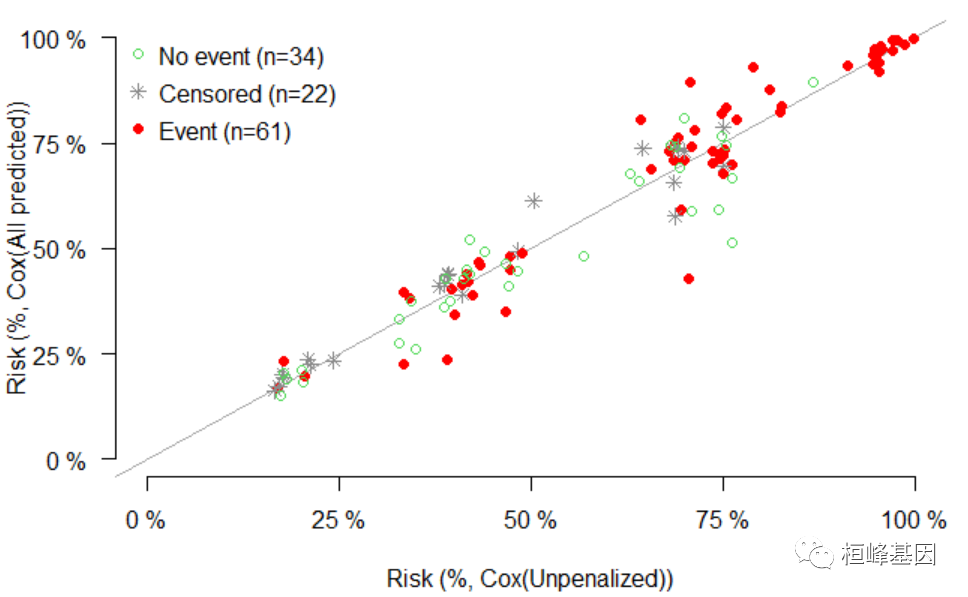
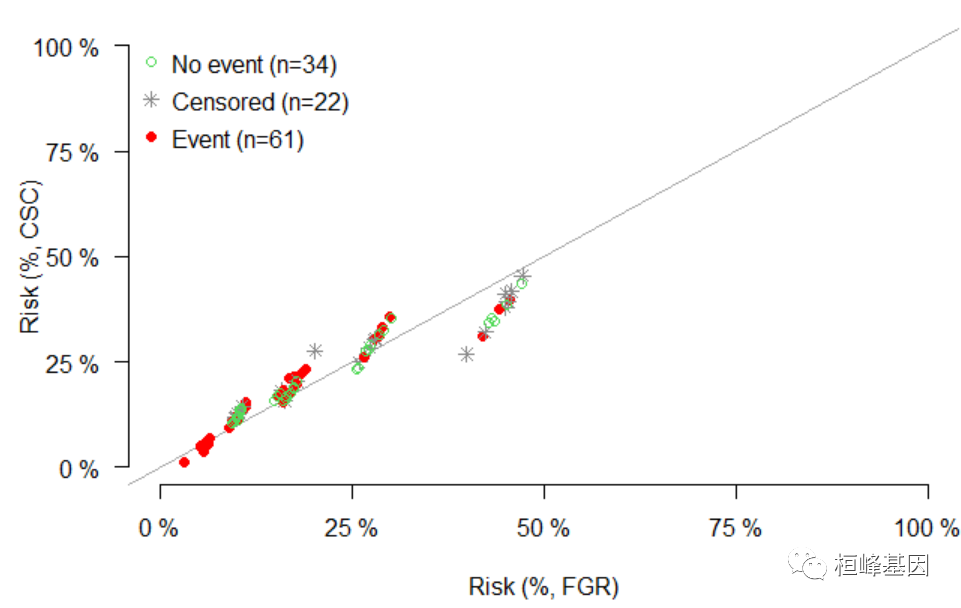
特异性Cox比例危险性回归
FGR: Formula interface for Fine-Gray regression competing risk models.
CSC: Interface for fitting cause-specific Cox proportional hazard regression models in competing risk.
## competing risk Not run: library(survival) m1 = FGR(Hist(time, status) ~ age + sex + ph.ecog + ph.karno, data = train_data, cause = 1) m2 = CSC(Hist(time, status) ~ age + sex + ph.ecog + ph.karno, data = train_data, cause = 1) xcr = Score(list(FGR = m1, CSC = m2), formula = Hist(time, status == 2) ~ 1, data = train_data, summary = "risks", null.model = 0L, times = c(365, 2 * 365)) plotRisk(xcr, times = c(365, 2 * 365))- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
整个过程是不是非常简单好用?学会了都是自己的本事,想学更多本领快来关注桓峰基因公众号,更多的免费文字版教程结合视频版教程相结合,保证您来了就有收获!!!
桓峰基因,铸造成功的您!
有想进生信交流群的老师可以扫最后一个二维码加微信,备注“单位+姓名+目的”,有些想发广告的就免打扰吧,还得费力气把你踢出去!
References:
-
Loprinzi CL. et al. Prospective evaluation of prognostic variables from patient-completed questionnaires. North Central Cancer Treatment Group. Journal of Clinical Oncology. 12(3):601-7, 1994.
-
Ulla B. Mogensen, Hemant Ishwaran, Thomas A. Gerds (2012). Evaluating Random Forests for Survival Analysis Using Prediction Error Curves. Journal of Statistical Software, 50(11), 1-23. DOI 10.18637/jss.v050.i11
-
TA Gerds, MW Kattan, M Schumacher, and C Yu. Estimating a time-dependent concordance index for survival prediction models with covariate dependent censoring. Statistics in Medicine, Ahead of print:to appear, 2013. DOI = 10.1002/sim.5681
-
Wolbers, M and Koller, MT and Witteman, JCM and Gerds, TA (2013) Concordance for prognostic models with competing risks Research report 13/3. Department of Biostatistics, University of Copenhagen
-
Andersen, PK (2012) A note on the decomposition of number of life years lost according to causes of death Research report 12/2. Department of Biostatistics, University of Copenhagen
-
Paul Blanche, Michael W Kattan, and Thomas A Gerds. The c-index is not proper for the evaluation of-year predicted risks. Biostatistics, 20(2): 347–357, 2018.
-
B. Ozenne, A. L. Soerensen, T.H. Scheike, C.T. Torp-Pedersen, and T.A. Gerds. riskregression: Predicting the risk of an event using Cox regression models. R Journal, 9(2):440–460, 2017.
-
J Benichou and Mitchell H Gail. Estimates of absolute cause-specific risk in cohort studies. Biometrics, pages 813–826, 1990.
-
T.A. Gerds, T.H. Scheike, and P.K. Andersen. Absolute risk regression for competing risks: Interpretation, link functions, and prediction. Statistics in Medicine, 31(29):3921–3930, 2012.
-
相关阅读:
旭日3SDB,安装原版ros
IDEA JRebel安装使用教程
【老生谈算法】matlab实现控制系统稳定性——控制系统
Preview(2021-2022年度第三届全国大学生算法设计与编程挑战赛(夏季赛)——正式赛——E)
web内容如何保护:如何有效地保护 HTML5 格式的视频内容?
抽丝剥茧:详述一次DevServer Proxy配置无效问题的细致排查过程
区块链交易平台服务器该怎么选
基于微信小程序的奶茶在线预定点单管理系统
代码随想录 Leetcode435. 无重叠区间
【每日一题Day345】LC2562找出数组的串联值 | 模拟
- 原文地址:https://blog.csdn.net/weixin_41368414/article/details/127347791
